4JG8
 
 | | Structure of RSK2 T493M CTD mutant bound to 2-cyano-N-(1-hydroxy-2-methylpropan-2-yl)-3-(3-(3,4,5-trimethoxyphenyl)-1H-indazol-5-yl)acrylamide | | Descriptor: | (2S)-2-cyano-N-(1-hydroxy-2-methylpropan-2-yl)-3-[3-(3,4,5-trimethoxyphenyl)-1H-indazol-5-yl]propanamide, Ribosomal protein S6 kinase alpha-3 | | Authors: | Miller, R.M, Paavilainen, V.O, Krishnan, S, Serafimova, I.M, Taunton, J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1002 Å) | | Cite: | Electrophilic fragment-based design of reversible covalent kinase inhibitors.
J.Am.Chem.Soc., 135, 2013
|
|
1FE2
 
 | | CRYSTAL STRUCTURE OF DIHOMO-GAMMA-LINOLEIC ACID BOUND IN THE CYCLOOXYGENASE CHANNEL OF PROSTAGLANDIN ENDOPEROXIDE H SYNTHASE-1. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EICOSA-8,11,14-TRIENOIC ACID, PROSTAGLANDIN ENDOPEROXIDE H SYNTHASE-1, ... | | Authors: | Thuresson, E.D, Malkowski, M.G, Lakkides, K.M, Smith, W.L, Garavito, R.M. | | Deposit date: | 2000-07-20 | | Release date: | 2001-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mutational and X-ray crystallographic analysis of the interaction of dihomo-gamma -linolenic acid with prostaglandin endoperoxide H synthases.
J.Biol.Chem., 276, 2001
|
|
4JNG
 
 | | Schmallenberg virus nucleoprotein-RNA complex | | Descriptor: | Nucleocapsid protein, RNA (42-MER) | | Authors: | Dong, H.H, Li, P, Bottcher, B, Elliott, R.M, Dong, C.J. | | Deposit date: | 2013-03-15 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of Schmallenberg orthobunyavirus nucleoprotein-RNA complex reveals a novel RNA sequestration mechanism.
Rna, 19, 2013
|
|
4JUK
 
 | | Crystal structure of H5N1 influenza virus hemagglutinin, clade 2.3.2.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2, ... | | Authors: | DuBois, R.M, Zaraket, H, Reddivari, M, Coop, T, Heath, R.J, White, S.W, Russell, C.J. | | Deposit date: | 2013-03-25 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7502 Å) | | Cite: | To be published
To be Published
|
|
4JUN
 
 | | Crystal structure of H5N1 influenza virus hemagglutinin, clade 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | DuBois, R.M, Zaraket, H, Reddivari, M, Coop, T, Heath, R.J, White, S.W, Russell, C.J. | | Deposit date: | 2013-03-25 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3398 Å) | | Cite: | To be published
To be Published
|
|
2G89
 
 | |
2G86
 
 | |
2GFA
 
 | | double tudor domain complex structure | | Descriptor: | Jumonji domain-containing protein 2A, peptide | | Authors: | Huang, Y, Fang, J, Bedford, M.T, Zhang, Y, Xu, R.M. | | Deposit date: | 2006-03-21 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A
Science, 312, 2006
|
|
2G8D
 
 | |
3D3T
 
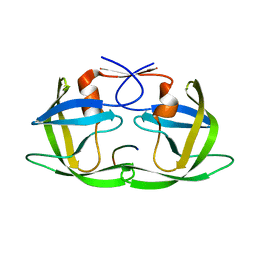 | | Crystal Structure of HIV-1 CRF01_AE in complex with the substrate p1-p6 | | Descriptor: | HIV-1 protease, P1-P6 SUBSTRATE PEPTIDE | | Authors: | Bandaranayake, R.M, Prabu-Jeyabalan, M, Kakizawa, J, Sugiura, W, Schiffer, C.A. | | Deposit date: | 2008-05-12 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of human immunodeficiency virus type 1 CRF01_AE protease in complex with the substrate p1-p6.
J.Virol., 82, 2008
|
|
4ASW
 
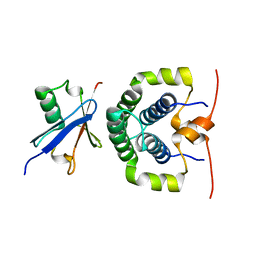 | | Structure of the complex between the N-terminal dimerisation domain of Sgt2 and the UBL domain of Get5 | | Descriptor: | SMALL GLUTAMINE-RICH TETRATRICOPEPTIDE REPEAT-CONTAINING PROTEIN 2, UBIQUITIN-LIKE PROTEIN MDY2 | | Authors: | Simon, A.C, Simpson, P.J, Goldstone, R.M, Krysztofinska, E.M, Murray, J.W, High, S, Isaacson, R.L. | | Deposit date: | 2012-05-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Sgt2/Get5 Complex Provides Insights Into Get-Mediated Targeting of Tail-Anchored Membrane Proteins
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4ASV
 
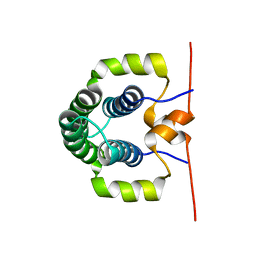 | | Solution structure of the N-terminal dimerisation domain of Sgt2 | | Descriptor: | SMALL GLUTAMINE-RICH TETRATRICOPEPTIDE REPEAT-CONTAINING PROTEIN 2 | | Authors: | Simon, A.C, Simpson, P.J, Goldstone, R.M, Krysztofinska, E.M, Murray, J.W, High, S, Isaacson, R.L. | | Deposit date: | 2012-05-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of the Sgt2/Get5 Complex Provides Insights Into Get-Mediated Targeting of Tail-Anchored Membrane Proteins
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5EQB
 
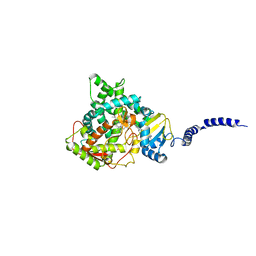 | | Crystal structure of lanosterol 14-alpha demethylase with intact transmembrane domain bound to itraconazole | | Descriptor: | 2-[(2R)-butan-2-yl]-4-{4-[4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazin-1-yl]phenyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Monk, B.C, Tomasiak, T.M, Keniya, M.V, Huschmann, F.U, Tyndall, J.D.A, O'Connell III, J.D, Cannon, R.D, Finer-Morre, J, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CY4
 
 | | The Cryo-Electron Microscopy Structure of the CorA channel from Methanocaldococcus jannaschii at 21.6 Angstrom in low magnesium. | | Descriptor: | MAGNESIUM TRANSPORT PROTEIN CORA | | Authors: | Cleverley, R.M, Kean, J, Shintre, C.A, Baldock, C, Derrick, J.P, Ford, R.C, Prince, S.M. | | Deposit date: | 2014-04-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (21.6 Å) | | Cite: | The Cryo-EM structure of the CorA channel from Methanocaldococcus jannaschii in low magnesium conditions.
Biochim. Biophys. Acta, 1848, 2015
|
|
5EQI
 
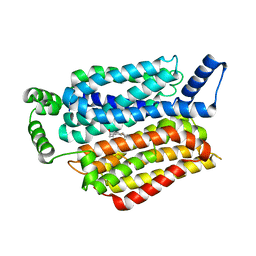 | | Human GLUT1 in complex with Cytochalasin B | | Descriptor: | Cytochalasin B, Solute carrier family 2, facilitated glucose transporter member 1 | | Authors: | Kapoor, K, Finer-Moore, J, Pedersen, B.P, Caboni, L, Waight, A.B, Hillig, R, Bringmann, P, Heisler, I, Muller, T, Siebeneicher, H, Stroud, R.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Mechanism of inhibition of human glucose transporter GLUT1 is conserved between cytochalasin B and phenylalanine amides.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4M9E
 
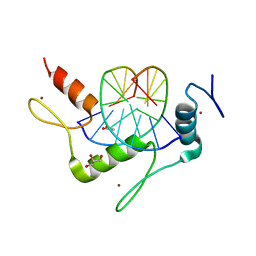 | | Structure of Klf4 zinc finger DNA binding domain in complex with methylated DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*GP*AP*GP*GP*(5CM)P*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*(5CM)P*GP*CP*CP*TP*C)-3'), ... | | Authors: | Liu, Y, Olanrewaju, Y.O, Blumenthal, R.M, Zhang, X, Cheng, X. | | Deposit date: | 2013-08-14 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for Klf4 recognition of methylated DNA.
Nucleic Acids Res., 42, 2014
|
|
1BO7
 
 | | THYMIDYLATE SYNTHASE R179T MUTANT | | Descriptor: | PROTEIN (THYMIDYLATE SYNTHASE), URIDINE-5'-MONOPHOSPHATE | | Authors: | Morse, R, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1998-08-10 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
2G8X
 
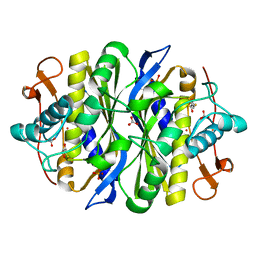 | | Escherichia coli Y209W apoprotein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CARBONATE ION, PHOSPHATE ION, ... | | Authors: | Lee, T.T, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2006-03-03 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The role of protein dynamics in thymidylate synthase catalysis: Variants of conserved dUMP-binding Tyr-261
TO BE PUBLISHED
|
|
1BP0
 
 | | THYMIDYLATE SYNTHASE R23I MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, POTASSIUM ION, PROTEIN (THYMIDYLATE SYNTHASE) | | Authors: | Morse, R, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1998-08-11 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
4KUI
 
 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain | | Descriptor: | ACETIC ACID, ISOPROPYL ALCOHOL, Regulatory protein SIR3 | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4KUD
 
 | | Crystal structure of N-terminal acetylated Sir3 BAH domain D205N mutant in complex with yeast nucleosome core particle | | Descriptor: | Histone H2A.2, Histone H2B.1, Histone H3, ... | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1BP6
 
 | | THYMIDYLATE SYNTHASE R23I, R179T DOUBLE MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, POTASSIUM ION, PROTEIN (THYMIDYLATE SYNTHASE) | | Authors: | Morse, R.J, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 1998-08-13 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
4BXS
 
 | | Crystal Structure of the Prothrombinase Complex from the Venom of Pseudonaja Textilis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lechtenberg, B.C, Murray-Rust, T.A, Johnson, D.J.D, Adams, T.E, Krishnaswamy, S, Camire, R.M, Huntington, J.A. | | Deposit date: | 2013-07-15 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Crystal Structure of the Prothrombinase Complex from the Venom of Pseudonaja Textilis.
Blood, 122, 2013
|
|
3B23
 
 | | Crystal structure of thrombin-variegin complex: Insights of a novel mechanism of inhibition and design of tunable thrombin inhibitors | | Descriptor: | Thrombin heavy chain, Thrombin light chain, Variegin | | Authors: | Koh, C.Y, Kumar, S, Swaminathan, K, Kini, R.M. | | Deposit date: | 2011-07-20 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of thrombin in complex with s-variegin: insights of a novel mechanism of inhibition and design of tunable thrombin inhibitors
Plos One, 6, 2011
|
|
5H3V
 
 | | Crystal structure of a Type IV Secretion System Component CagX in Helicobacter pylori | | Descriptor: | Cag8, DI(HYDROXYETHYL)ETHER, ISOPROPYL ALCOHOL | | Authors: | Zhang, J, Wu, Y, Zhao, Y, Sun, L, Keegan, R.M, Liu, Y, Isupov, M.N. | | Deposit date: | 2016-10-27 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the type IV secretion system component CagX from Helicobacter pylori
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
