1W2D
 
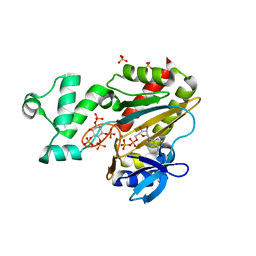 | | Human Inositol (1,4,5)-trisphosphate 3-kinase complexed with Mn2+/ADP/Ins(1,3,4,5)P4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, INOSITOL-TRISPHOSPHATE 3-KINASE A, ... | | Authors: | Gonzalez, B, Schell, M.J, Irvine, R.F, Williams, R.L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure of a Human Inositol 1,4,5-Trisphosphate 3-Kinase; Substrate Binding Reveals Why It is not a Phosphoinositide 3-Kinase
Mol.Cell, 15, 2004
|
|
1W72
 
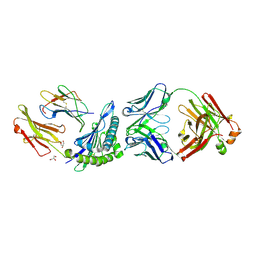 | | Crystal structure of HLA-A1:MAGE-A1 in complex with Fab-Hyb3 | | Descriptor: | BETA-2-MICROGLOBULIN, GLYCEROL, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Hulsmeyer, M, Chames, P, Hillig, R.C, Stanfield, R.L, Held, G, Coulie, P.G, Alings, C, Wille, G, Saenger, W, Uchanska-Ziegler, B, Hoogenboom, H.R, Ziegler, A. | | Deposit date: | 2004-08-27 | | Release date: | 2004-11-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Major Histocompatibility Complex.Peptide- Restricted Antibody and T Cell Receptor Molecules Recognize Their Target by Distinct Binding Modes: Crystal Structure of Human Leukocyte Antigen (Hla)-A1.Mage-A1 in Complex with Fab-Hyb3
J.Biol.Chem., 280, 2005
|
|
1WAC
 
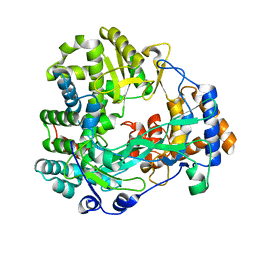 | | Back-priming mode of Phi6 RNA-dependent RNA polymerase | | Descriptor: | P2 PROTEIN | | Authors: | Laurila, M.R.L, Salgado, P.S, Stuart, D.I, Grimes, J.M, Bamford, D.H. | | Deposit date: | 2004-10-26 | | Release date: | 2005-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Back-Priming Mode of Phi6 RNA-Dependent RNA Polymerase
J.Gen.Virol., 86, 2005
|
|
1W7P
 
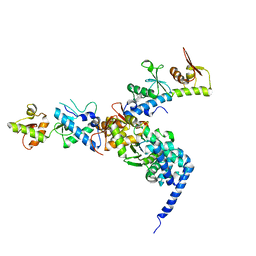 | | The crystal structure of endosomal complex ESCRT-II (VPS22/VPS25/VPS36) | | Descriptor: | VPS22, YPL002C, VPS25, ... | | Authors: | Teo, H, Perisic, O, Gonzalez, B, Williams, R.L. | | Deposit date: | 2004-09-07 | | Release date: | 2004-09-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Escrt-II, an Endosome-Associated Complex Required for Protein Sorting: Crystal Structure and Interactions with Escrt-III and Membranes
Dev.Cell, 7, 2004
|
|
1WKP
 
 | | Flowering locus t (ft) from arabidopsis thaliana | | Descriptor: | FLOWERING LOCUS T protein, SULFATE ION | | Authors: | Miller, D, Banfield, M.J, Winter, V.J, Brady, R.L. | | Deposit date: | 2004-06-01 | | Release date: | 2005-06-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A divergent external loop confers antagonistic activity on floral regulators FT and TFL1.
Embo J., 25, 2006
|
|
1TT2
 
 | | Cryogenic crystal structure of Staphylococcal nuclease variant truncated Delta+PHS I92K | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Nguyen, D.M, Reynald, R.L, Gittis, A.G, Lattman, E.E. | | Deposit date: | 2004-06-21 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray and thermodynamic studies of staphylococcal nuclease variants I92E and I92K: insights into polarity of the protein interior
J.Mol.Biol., 341, 2004
|
|
1UPT
 
 | | Structure of a complex of the golgin-245 GRIP domain with Arl1 | | Descriptor: | ADP-RIBOSYLATION FACTOR-LIKE PROTEIN 1, GOLGI AUTOANTIGEN, GOLGIN SUBFAMILY A MEMBER 4, ... | | Authors: | Panic, B, Perisic, O, Veprintsev, D.B, Williams, R.L, Munro, S. | | Deposit date: | 2003-10-12 | | Release date: | 2003-10-30 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Arl1-Dependent Targeting of Homodimeric Grip Domains to the Golgi Apparatus
Mol.Cell, 12, 2003
|
|
1TTM
 
 | | Human carbonic anhydrase II complexed with 667-coumate | | Descriptor: | 6-OXO-8,9,10,11-TETRAHYDRO-7H-CYCLOHEPTA[C][1]BENZOPYRAN-3-O-SULFAMATE, Carbonic anhydrase II, ZINC ION | | Authors: | Lloyd, M.D, Pederick, R.L, Natesh, R, Woo, L.W.L, Purohit, A, Reed, M.J, Acharya, K.R, Potter, B.V.L. | | Deposit date: | 2004-06-23 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human carbonic anhydrase II at 1.95 A resolution in complex with 667-coumate, a novel anti-cancer agent
Biochem.J., 385, 2005
|
|
1XRU
 
 | |
1XIV
 
 | | Plasmodium falciparum lactate dehydrogenase complexed with 2-({4-chloro-[hydroxy(methoxy)methyl]cyclohexyl}amino)ethane-1,1,2-triol | | Descriptor: | 2-({4-CHLORO-2-[HYDROXY(METHOXY)METHYL]CYCLOHEXYL}AMINO)ETHANE-1,1,2-TRIOL, GLYCEROL, L-lactate dehydrogenase | | Authors: | Conners, R, Cameron, A, Read, J.A, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-09-22 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium
falciparum lactate dehydrogenase
MOL.BIOCHEM.PARASITOL., 142, 2005
|
|
1TIH
 
 | |
1XME
 
 | | Structure of Recombinant Cytochrome ba3 Oxidase from Thermus thermophilus | | Descriptor: | COPPER (II) ION, Cytochrome c oxidase polypeptide I, Cytochrome c oxidase polypeptide II, ... | | Authors: | Hunsicker-Wang, L.M, Pacoma, R.L, Chen, Y, Fee, J.A, Stout, C.D. | | Deposit date: | 2004-10-01 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel cryoprotection scheme for enhancing the diffraction of crystals of recombinant cytochrome ba3 oxidase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1U4S
 
 | | Plasmodium falciparum lactate dehydrogenase complexed with 2,6-naphthalenedisulphonic acid | | Descriptor: | L-lactate dehydrogenase, NAPHTHALENE-2,6-DISULFONIC ACID | | Authors: | Conners, R, Cameron, A, Read, J, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-07-26 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium falciparum lactate dehydrogenase.
Mol.Biochem.Parasitol., 142, 2005
|
|
1U21
 
 | | transthyretin with tethered inhibitor on one monomer. | | Descriptor: | 2-[(3,5-DICHLORO-4-TRIOXIDANYLPHENYL)AMINO]BENZOIC ACID, Transthyretin | | Authors: | Wiseman, R.L, Johnson, S.M, Kelker, M.S, Foss, T, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2004-07-16 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Kinetic stabilization of an oligomeric protein by a single ligand binding event
J.Am.Chem.Soc., 127, 2005
|
|
1U4O
 
 | | Plasmodium falciparum lactate dehydrogenase complexed with 2,6-naphthalenedicarboxylic acid | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,6-DICARBOXYNAPHTHALENE, L-lactate dehydrogenase | | Authors: | Conners, R, Cameron, A, Read, J, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-07-26 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium falciparum lactate dehydrogenase.
Mol.Biochem.Parasitol., 142, 2005
|
|
1U5C
 
 | | Plasmodium falciparum lactate dehydrogenase complexed with 3,7-dihydroxynaphthalene-2-carboxylic acid and NAD+ | | Descriptor: | 3,7-DIHYDROXY-2-NAPHTHOIC ACID, L-lactate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Conners, R, Cameron, A, Read, J, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-07-27 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium falciparum lactate dehydrogenase.
Mol.Biochem.Parasitol., 142, 2005
|
|
1XQ8
 
 | |
1XZN
 
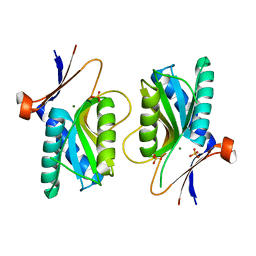 | | PYRR, THE REGULATOR OF THE PYRIMIDINE BIOSYNTHETIC OPERON IN BACILLUS CALDOLYTICUS, sulfate-bound form | | Descriptor: | MAGNESIUM ION, PyrR bifunctional protein, SULFATE ION | | Authors: | Chander, P, Halbig, K.M, Miller, J.K, Fields, C.J, Bonner, H.K, Grabner, G.K, Switzer, R.L, Smith, J.L. | | Deposit date: | 2004-11-12 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of the Nucleotide Complex of PyrR, the pyr Attenuation Protein from Bacillus caldolyticus, Suggests Dual Regulation by Pyrimidine and Purine Nucleotides.
J.Bacteriol., 187, 2005
|
|
1XNC
 
 | |
1XZ8
 
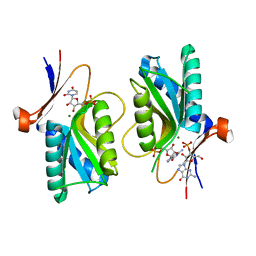 | | Pyrr, The Regulator Of The Pyrimidine Biosynthetic Operon In Bacillus caldolyticus, Nucleotide-bound form | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chander, P, Halbig, K.M, Miller, J.K, Fields, C.J, Bonner, H.K, Grabner, G.K, Switzer, R.L, Smith, J.L. | | Deposit date: | 2004-11-11 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Nucleotide Complex of PyrR, the pyr Attenuation Protein from Bacillus caldolyticus, Suggests Dual Regulation by Pyrimidine and Purine Nucleotides.
J.Bacteriol., 187, 2005
|
|
5DFZ
 
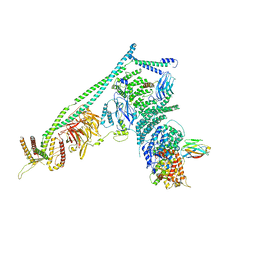 | | Structure of Vps34 complex II from S. cerevisiae. | | Descriptor: | Nanobody binding S. cerevisiae Vps34, Phosphatidylinositol 3-kinase VPS34, Putative N-terminal domain of S. cerevisiae Vps30, ... | | Authors: | Rostislavleva, K, Soler, N, Ohashi, Y, Zhang, L, Williams, R.L. | | Deposit date: | 2015-08-27 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes.
Science, 350, 2015
|
|
5DHG
 
 | | The crystal structure of nociceptin/orphanin FQ peptide receptor (NOP) in complex with C-35 (PSI Community Target) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-benzyl-N-{3-[4-(2,6-dichlorophenyl)piperidin-1-yl]propyl}-D-prolinamide, OLEIC ACID, ... | | Authors: | Miller, R.L, Thompson, A.A, Trapella, C, Guerrini, R, Malfacini, D, Patel, N, Han, G.W, Cherezov, V, Calo, G, Katritch, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2015-08-30 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Importance of Ligand-Receptor Conformational Pairs in Stabilization: Spotlight on the N/OFQ G Protein-Coupled Receptor.
Structure, 23, 2015
|
|
1YNV
 
 | | Asp79 makes a large, unfavorable contribution to the stability of RNase Sa | | Descriptor: | Guanyl-specific ribonuclease Sa, SULFATE ION | | Authors: | Trevino, S.R, Gokulan, K, Newsom, S, Thurlkill, R.L, Shaw, K.L, Mitkevich, V.A, Makarov, A.A, Sacchettini, J.C, Scholtz, J.M, Pace, C.N. | | Deposit date: | 2005-01-25 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Asp79 Makes a Large, Unfavorable Contribution to the Stability of RNase Sa.
J.Mol.Biol., 354, 2005
|
|
3NZP
 
 | | Crystal Structure of the Biosynthetic Arginine decarboxylase SpeA from Campylobacter jejuni, Northeast Structural Genomics Consortium Target BR53 | | Descriptor: | Arginine decarboxylase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-07-16 | | Release date: | 2010-09-01 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of bacterial biosynthetic arginine decarboxylases.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3NN8
 
 | |
