4W7U
 
 | |
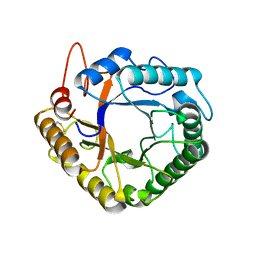
4W7W
 
 | |
4W7V
 
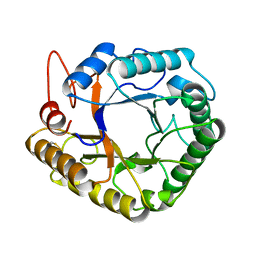 | |
4M1R
 
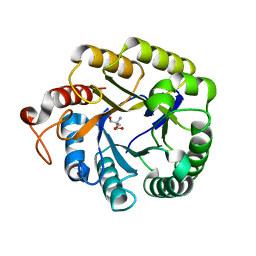 | | Structure of a novel cellulase 5 from a sugarcane soil metagenomic library | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellulase 5 | | Authors: | Paiva, J.H, Alvarez, T.M, Cairo, J.P, Paixao, D.A, Almeida, R.A, Tonoli, C.C.C, Ruiz, D.M, Ruller, R, Santos, C.R, Squina, F.M, Murakami, M.T. | | Deposit date: | 2013-08-03 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of a novel cellulase 5 from sugarcane soil metagenome.
Plos One, 8, 2013
|
|
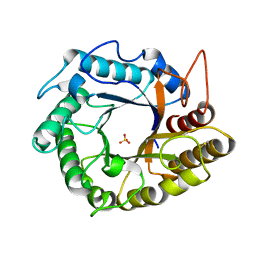
5HOS
 
 | |
5HPC
 
 | |
5HNN
 
 | |
2L8A
 
 | | Structure of a novel CBM3 lacking the calcium-binding site | | Descriptor: | Endoglucanase | | Authors: | Paiva, J.H, Meza, A.N, Sforca, M.L, Navarro, R.Z, Neves, J.L, Santos, C.R, Murakami, M.T, Zeri, A.C. | | Deposit date: | 2011-01-07 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dissecting structure-function-stability relationships of a thermostable GH5-CBM3 cellulase from Bacillus subtilis 168.
Biochem.J., 441, 2012
|
|
3NN8
 
 | |
4NKM
 
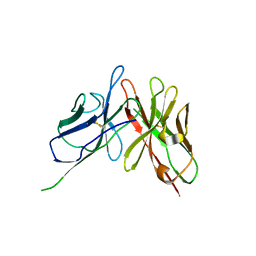 | | Crystal structure of engineered anti-EE scFv antibody fragment | | Descriptor: | Engineered scFv | | Authors: | Kalyoncu, S, Hyun, J, Pai, J.C, Johnson, J.L, Etzminger, K, Jain, A, Heaner Jr, D, Molares, I.A, Truskett, T.M, Maynard, J.A, Lieberman, R.L. | | Deposit date: | 2013-11-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Effects of protein engineering and rational mutagenesis on crystal lattice of single chain antibody fragments.
Proteins, 82, 2014
|
|
4NKD
 
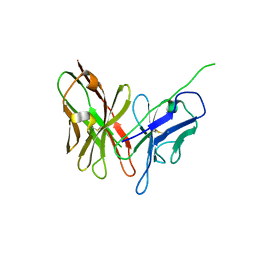 | | Crystal structure of engineered anti-EE scFv antibody fragment | | Descriptor: | Engineered scFv | | Authors: | Kalyoncu, S, Hyun, J, Pai, J.C, Johnson, J.L, Etzminger, K, Jain, A, Heaner Jr, D, Molares, I.A, Truskett, T.M, Maynard, J.A, Lieberman, R.L. | | Deposit date: | 2013-11-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Effects of protein engineering and rational mutagenesis on crystal lattice of single chain antibody fragments.
Proteins, 82, 2014
|
|
4NKO
 
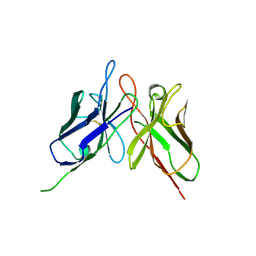 | | Crystal structure of engineered anti-EE scFv antibody fragment | | Descriptor: | Engineered scFv | | Authors: | Kalyoncu, S, Hyun, J, Pai, J.C, Johnson, J.L, Etzminger, K, Jain, A, Heaner Jr, D, Molares, I.A, Truskett, T.M, Maynard, J.A, Lieberman, R.L. | | Deposit date: | 2013-11-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.496 Å) | | Cite: | Effects of protein engineering and rational mutagenesis on crystal lattice of single chain antibody fragments.
Proteins, 82, 2014
|
|
2FD0
 
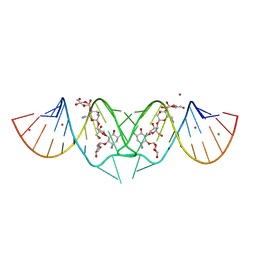 | | HIV-1 DIS kissing-loop in complex with lividomycin | | Descriptor: | (2R,3S,4S,5S,6R)-2-((2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3S,4R,5S)-5-((1R,2R,3S,5R,6S)-3,5-DIAMINO-2-((2S,3R ,5S,6R)-3-AMINO-5-HYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-4-HYDROXY-2-(HYDROXYMET HYL)-TETRAHYDROFURAN-3-YLOXY)-4-HYDROXY-TETRAHYDRO-2H-PYRAN-3-YLOXY)-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-3,4,5-TRIOL, CHLORIDE ION, HIV-1 DIS RNA, ... | | Authors: | Ennifar, E, Paillart, J.C, Marquet, R, Dumas, P. | | Deposit date: | 2005-12-13 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: from crystal to cell.
Nucleic Acids Res., 34, 2006
|
|
2FCX
 
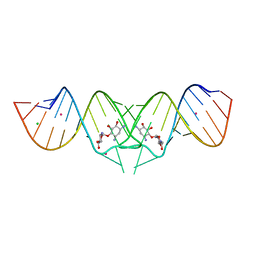 | | HIV-1 DIS kissing-loop in complex with neamine | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, CHLORIDE ION, HIV-1 DIS RNA, ... | | Authors: | Ennifar, E, Paillart, J.C, Marquet, R, Dumas, P. | | Deposit date: | 2005-12-13 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: from crystal to cell.
Nucleic Acids Res., 34, 2006
|
|
5CG9
 
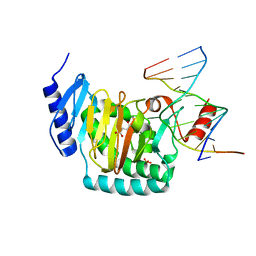 | | NgTET1 in complex with 5mC DNA in space group P3221 | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, DNA (5'-D(*TP*GP*TP*CP*AP*GP*(5CM)P*GP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Hashimoto, H, Pais, J.E, Dai, N, Zhang, X, Zheng, Y, Cheng, X. | | Deposit date: | 2015-07-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Structure of Naegleria Tet-like dioxygenase (NgTet1) in complexes with a reaction intermediate 5-hydroxymethylcytosine DNA.
Nucleic Acids Res., 43, 2015
|
|
5CG8
 
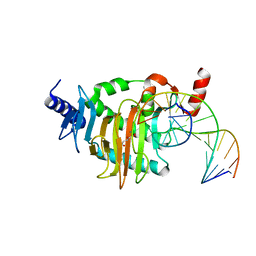 | | NgTET1 in complex with 5hmC DNA | | Descriptor: | 2-OXOGLUTARIC ACID, DNA (5'-D(*AP*GP*AP*AP*TP*TP*CP*CP*GP*TP*TP*CP*CP*A)-3'), DNA (5'-D(*TP*GP*GP*AP*AP*(5HC)P*GP*GP*AP*AP*TP*TP*CP*T)-3'), ... | | Authors: | Hashimoto, H, Pais, J.E, Dai, N, Zhang, X, Zheng, Y, Cheng, X. | | Deposit date: | 2015-07-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structure of Naegleria Tet-like dioxygenase (NgTet1) in complexes with a reaction intermediate 5-hydroxymethylcytosine DNA.
Nucleic Acids Res., 43, 2015
|
|
4LT5
 
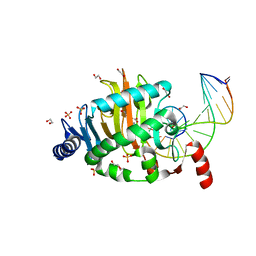 | | Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA, MANGANESE (II) ION, ... | | Authors: | Hashimoto, H, Pais, J.E, Zhang, X, Saleh, L, Fu, Z.Q, Dai, N, Correa, I.R, Roberts, R.J, Zheng, Y, Cheng, X. | | Deposit date: | 2013-07-23 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA.
Nature, 506, 2014
|
|
7AER
 
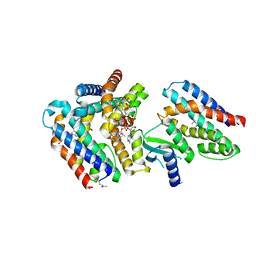 | | Rebuilt and re-refined PDB entry 5yep: tri-AMPylated Shewanella oneidensis HEPN toxin in complex with MNT antitoxin | | Descriptor: | ADENOSINE MONOPHOSPHATE, Toxin-antitoxin system antidote Mnt family, Toxin-antitoxin system toxin HepN family | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-18 | | Release date: | 2020-12-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
7AE9
 
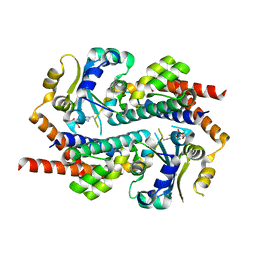 | | Crystal structure of mono-AMPylated HEPN(R46E) toxin in complex with MNT antitoxin | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-MONOPHOSPHATE, HEPN toxin, MNT ANTITOXIN | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
7AE6
 
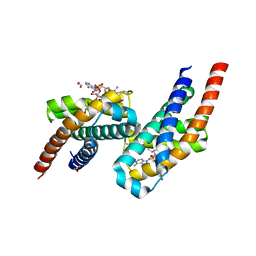 | | Crystal structure of di-AMPylated HEPN(R102A) toxin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, HEPN toxin | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
7AE8
 
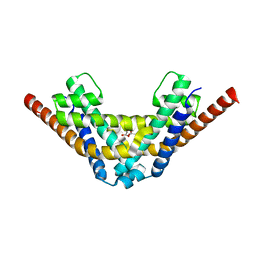 | | Crystal structure of HEPN(R102A) toxin | | Descriptor: | ACETATE ION, HEPN toxin | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
7AE2
 
 | | Crystal structure of HEPN(H107A-Y109F) toxin in complex with MNT antitoxin | | Descriptor: | HEPN toxin, MNT ANTITOXIN | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-17 | | Release date: | 2020-12-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
4X2Q
 
 | | Crystal Structure of Human Aldehyde Dehydrogenase, ALDH1a2 | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Retinal dehydrogenase 2 | | Authors: | Stenkamp, R.E, Le Trong, I, Amory, J.K, Paik, J, Goldstein, A.S. | | Deposit date: | 2014-11-26 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Synthesis and In Vitro Testing of Bisdichloroacetyldiamine Analogs for Use as a Reversible Male Contraceptive
To Be Published
|
|
6UFV
 
 | |
6UFW
 
 | |
