6COG
 
 | | AtHNL enantioselectivity mutant At-A9-H7 Apo, Y13C,Y121L,P126F,L128W,C131T,A209I with benzaldehyde | | Descriptor: | Alpha-hydroxynitrile lyase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jones, B.J, Kazlauskas, R.J, Desrouleaux, R. | | Deposit date: | 2018-03-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | AtHNL enantioselectivity mutants
To be published
|
|
6CED
 
 | | Crystal structure of fragment 3-(3-Methyl-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(3-methyl-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Franzoni, I, Ravichandran, M, Lautens, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
6CWI
 
 | |
4NX8
 
 | | Structure of a PTP-like phytase from Bdellovibrio bacteriovorus | | Descriptor: | GLYCEROL, MAGNESIUM ION, Protein-tyrosine phosphatase 2 | | Authors: | Gruninger, R.J, Lovering, A.L. | | Deposit date: | 2013-12-08 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Structural and biochemical analysis of a unique phosphatase from Bdellovibrio bacteriovorus reveals its structural and functional relationship with the protein tyrosine phosphatase class of phytase.
Plos One, 9, 2014
|
|
6COD
 
 | | AtHNL enantioselectivity mutant At-A9-H7 Apo Y13C,Y121L,P126F,L128W,C131T,F179L,A209I with benzaldehyde | | Descriptor: | Alpha-hydroxynitrile lyase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jones, B.J, Kazlauskas, R.J, Desrouleaux, R. | | Deposit date: | 2018-03-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | AtHNL enantioselectivity mutants
To be published
|
|
6COE
 
 | | AtHNL enantioselectivity mutant At-A9-H7 Apo Y13C,Y121L,P126F,L128W,C131T,F179L,A209I with benzaldehyde, MANDELIC ACID NITRILE | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, (S)-MANDELIC ACID NITRILE, Alpha-hydroxynitrile lyase, ... | | Authors: | Jones, B.J, Kazlauskas, R.J, Desrouleaux, R. | | Deposit date: | 2018-03-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | AtHNL enantioselectivity mutants
To be published
|
|
7T4J
 
 | | Crystal Structure of EGFR_D770_N771insNPG/V948R in complex with TAK-788 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Epidermal growth factor receptor, ... | | Authors: | Skene, R.J, Lane, W, Hu, Y. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of mobocertinib, a potent, oral inhibitor of EGFR exon 20 insertion mutations in non-small cell lung cancer.
Bioorg.Med.Chem.Lett., 80, 2022
|
|
7T91
 
 | |
6CWL
 
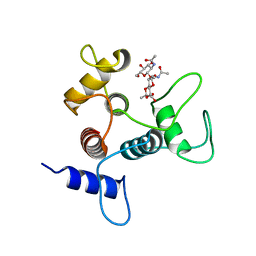 | | Crystal structure of SpaA-SLH in complex with beta-D-GlcNAc-(1->3)-4,6-Pyr-beta-D-ManNAcOMe | | Descriptor: | (2S,4aR,6R,7S,8R,8aS)-7-(acetylamino)-6-({2-(acetylamino)-3-O-[2-(acetylamino)-2-deoxy-beta-D-glucopyranosyl]-4,6-O-[(1S)-1-carboxylic acidethylidene]-2-deoxy-beta-D-mannopyranosyl}oxy)-8-{[2-(acetylamino)-2-deoxy-beta-D-glucopyranosyl]oxy}-2-methylhexahydro-2H-pyrano[3,2-d][1,3]dioxine-2-carboxylic acid, Surface (S-) layer glycoprotein | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2018-03-30 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of cell wall anchoring by SLH domains in Paenibacillus alvei.
Nat Commun, 9, 2018
|
|
7T4I
 
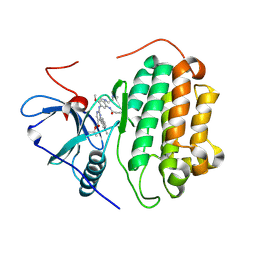 | | Crystal Structure of wild type EGFR in complex with TAK-788 | | Descriptor: | Epidermal growth factor receptor, propan-2-yl 2-[[4-[2-(dimethylamino)ethyl-methyl-amino]-2-methoxy-5-(propanoylamino)phenyl]amino]-4-(1-methylindol-3-yl)pyrimidine-5-carboxylate | | Authors: | Skene, R.J, Lane, W. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of mobocertinib, a potent, oral inhibitor of EGFR exon 20 insertion mutations in non-small cell lung cancer.
Bioorg.Med.Chem.Lett., 80, 2022
|
|
7T9P
 
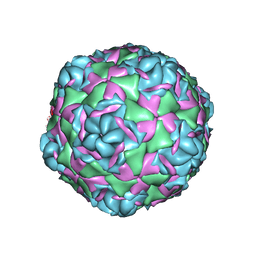 | |
7TAH
 
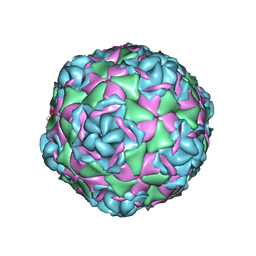 | |
7TAJ
 
 | |
7TAF
 
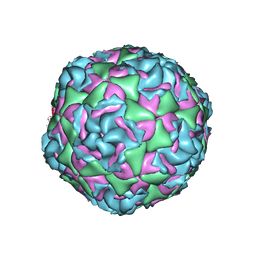 | | Cryo-EM structure of Human Enterovirus D68 US/MO/14-18947 strain virion in complex with inhibitor 11526092 | | Descriptor: | N,N-dimethyl-5-(3-{2-methyl-4-[5-(trifluoromethyl)-1,2,4-oxadiazol-3-yl]phenoxy}propyl)-1,2-oxazole-3-carboxamide, viral protein 1, viral protein 2, ... | | Authors: | Fu, J, Klose, T, Kuhn, R.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-20 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Isoxazole-3-Carboxamide Derivatives of Pleconaril Destabilize the Viral Capsid of Enterovirus-D68
To Be Published
|
|
7TAG
 
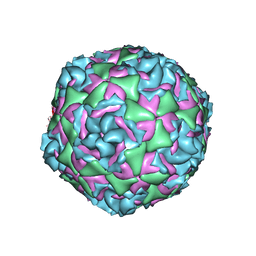 | | Cryo-EM structure of Human Enterovirus D68 US/MO/14-18947 strain virion in complex with pleconaril | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, viral protein 1, viral protein 2, ... | | Authors: | Fu, J, Klose, T, Kuhn, R.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-20 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Isoxazole-3-Carboxamide Derivatives of Pleconaril Destabilize the Viral Capsid of Enterovirus-D68
To Be Published
|
|
4OX5
 
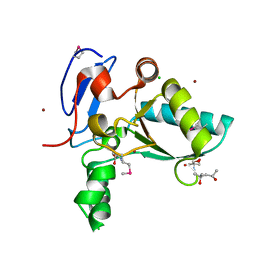 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-02-04 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
6CWC
 
 | | Crystal structure of SpaA-SLH | | Descriptor: | CHLORIDE ION, SULFATE ION, Surface (S-) layer glycoprotein | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2018-03-30 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of cell wall anchoring by SLH domains in Paenibacillus alvei.
Nat Commun, 9, 2018
|
|
4OXD
 
 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | Descriptor: | CHLORIDE ION, LYSINE, LdcB LD-carboxypeptidase, ... | | Authors: | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-02-05 | | Release date: | 2014-05-21 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
6CGX
 
 | | Backbone cyclised conotoxin Vc1.1 mutant - D11A, E14A | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Clark, R.J. | | Deposit date: | 2018-02-21 | | Release date: | 2018-05-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Studies Reveal the Molecular Basis for GABAB-Receptor Mediated Inhibition of High Voltage-Activated Calcium Channels by alpha-Conotoxin Vc1.1.
ACS Chem. Biol., 13, 2018
|
|
4AYM
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 3 P106A mutant | | Descriptor: | COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
4OZT
 
 | | crystal structure of the ligand binding domains of the Bovicola ovis ecdysone receptor EcR/USP heterodimer (PonA crystal) | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone receptor, N-ETHYLMALEIMIDE, ... | | Authors: | Ren, B, Peat, T.S, Streltsov, V.A, Pollard, M, Fernley, R, Grusovin, J, Seabrook, S, Pilling, P, Phan, T, Lu, L, Lovrecz, G.O, Graham, L.D, Hill, R.J. | | Deposit date: | 2014-02-19 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unprecedented conformational flexibility revealed in the ligand-binding domains of the Bovicola ovis ecdysone receptor (EcR) and ultraspiracle (USP) subunits.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AYD
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 1 R106A mutant | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules
Plos Pathog., 8, 2012
|
|
4OZR
 
 | | Crystal structure of the ligand binding domains of the Bovicola ovis ecdysone receptor EcR/USP heterodimer (methylene lactam crystal) | | Descriptor: | Ecdysone receptor, Retinoid X receptor | | Authors: | Ren, B, Peat, T.S, Streltsov, V.A, Pollard, M, Fernley, R, Grusovin, J, Seabrook, S, Pilling, P, Phan, T, Lu, L, Lovrecz, G.O, Graham, L.D, Hill, R.J. | | Deposit date: | 2014-02-18 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unprecedented conformational flexibility revealed in the ligand-binding domains of the Bovicola ovis ecdysone receptor (EcR) and ultraspiracle (USP) subunits.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6DDA
 
 | | Nurr1 Covalently Modified by a Dopamine Metabolite | | Descriptor: | 5-hydroxy-1,2-dihydro-6H-indol-6-one, BROMIDE ION, Nuclear receptor subfamily 4 group A member 2, ... | | Authors: | Bruning, J.M, Wang, Y, Otrabella, F, Boxue, T, Liu, H, Bhattacharya, P, Guo, S, Holton, J.M, Fletterick, R.J, Jacobson, M.P, England, P.M. | | Deposit date: | 2018-05-09 | | Release date: | 2019-03-20 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Covalent Modification and Regulation of the Nuclear Receptor Nurr1 by a Dopamine Metabolite.
Cell Chem Biol, 26, 2019
|
|
6DO1
 
 | | Structure of nanobody-stabilized angiotensin II type 1 receptor bound to S1I8 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-like peptide S1I8, ... | | Authors: | Wingler, L.M, McMahon, C, Staus, D.P, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2018-06-08 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Distinctive Activation Mechanism for Angiotensin Receptor Revealed by a Synthetic Nanobody.
Cell, 176, 2019
|
|
