4ZA1
 
 | | Crystal Structure of NosA Involved in Nosiheptide Biosynthesis | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NosA | | Authors: | Liu, S, Guo, H, Zhang, T, Han, L, Yao, P, Zhang, Y, Rong, N, Yu, Y, Lan, W, Wang, C, Ding, J, Wang, R, Liu, W, Cao, C. | | Deposit date: | 2015-04-13 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based Mechanistic Insights into Terminal Amide Synthase in Nosiheptide-Represented Thiopeptides Biosynthesis
Sci Rep, 5, 2015
|
|
1DE3
 
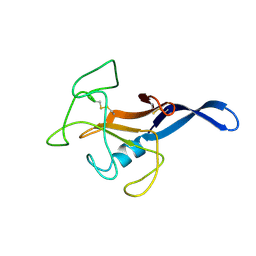 | | SOLUTION STRUCTURE OF THE CYTOTOXIC RIBONUCLEASE ALPHA-SARCIN | | Descriptor: | RIBONUCLEASE ALPHA-SARCIN | | Authors: | Perez-Canadillas, J.M, Campos-Olivas, R, Santoro, J, Lacadena, J, Martinez del Pozo, A, Gavilanes, J.G, Rico, M, Bruix, M. | | Deposit date: | 1999-11-12 | | Release date: | 2000-06-21 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The highly refined solution structure of the cytotoxic ribonuclease alpha-sarcin reveals the structural requirements for substrate recognition and ribonucleolytic activity.
J.Mol.Biol., 299, 2000
|
|
4BCB
 
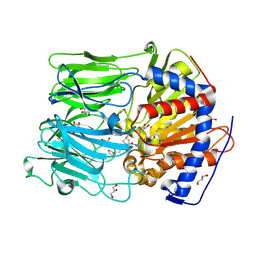 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN WITH A COVALENTLY BOUND P2- substituted N-acyl-prolylpyrrolidine inhibitor | | Descriptor: | (5R,6R,8S)-8-(3-{[AMINO(IMINO)METHYL]AMINO}PHENYL)-5-CYCLOHEXYL-6-HYDROXY-3-OXO-1-PHENYL-2,7-DIOXA-4-AZA-6-PHOSPHANONAN-9-OIC ACID 6-OXIDE, GLYCEROL, PROLYL ENDOPEPTIDASE, ... | | Authors: | VanDerVeken, P, Fulop, V, Rea, D, Gerard, M, VanElzen, R, Joossens, J, Cheng, J.D, Baekelandt, V, DeMeester, I, Lambeir, A.M, Augustyns, K. | | Deposit date: | 2012-10-01 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P2-Substituted N-Acylprolylpyrrolidine Inhibitors of Prolyl Oligopeptidase: Biochemical Evaluation, Binding Mode Determination, and Assessment in a Cellular Model of Synucleinopathy.
J.Med.Chem., 55, 2012
|
|
4ZNS
 
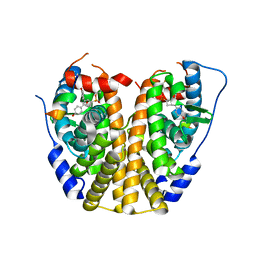 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 3-Fluoro-substituted OBHS derivative | | Descriptor: | 3-fluorophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
4ZO2
 
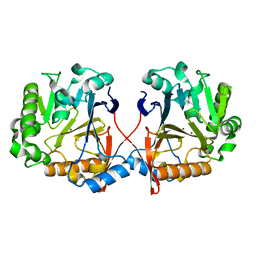 | | AidC, a Dizinc Quorum-Quenching Lactonase | | Descriptor: | Acylhomoserine lactonase, ZINC ION | | Authors: | Mascarenhas, R, Thomas, P.W, Wu, C.-X, Nocek, B.P, Hoang, Q, Fast, W, Liu, D. | | Deposit date: | 2015-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Structural and Biochemical Characterization of AidC, a Quorum-Quenching Lactonase with Atypical Selectivity.
Biochemistry, 54, 2015
|
|
1HLG
 
 | | CRYSTAL STRUCTURE OF HUMAN GASTRIC LIPASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LIPASE, ... | | Authors: | Roussel, A, Canaan, S, Verger, R, Cambillau, C. | | Deposit date: | 1999-03-12 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human gastric lipase and model of lysosomal acid lipase, two lipolytic enzymes of medical interest.
J.Biol.Chem., 274, 1999
|
|
4ZOL
 
 | | Crystal Structure of Tubulin-Stathmin-TTL-Tubulysin M Complex | | Descriptor: | (2R,4R)-4-{[(2-{(1R,3R)-1-(acetyloxy)-4-methyl-3-[methyl(N-{[(2S)-1-methylpiperidin-2-yl]carbonyl}-D-isoleucyl)amino]pentyl}-1,3-thiazol-4-yl)carbonyl]amino}-2-methyl-5-phenylpentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Zhang, R. | | Deposit date: | 2015-05-06 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules
Mol.Pharmacol., 89, 2016
|
|
1HJM
 
 | | HUMAN PRION PROTEIN AT PH 7.0 | | Descriptor: | MAJOR PRION PROTEIN PRECURSOR | | Authors: | Calzolai, L, Zahn, R. | | Deposit date: | 2003-02-27 | | Release date: | 2003-07-03 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Influence of Ph on NMR Structure and Stability of the Human Prion Protein Globular Domain
J.Biol.Chem., 278, 2003
|
|
1HP0
 
 | | CRYSTAL STRUCTURE OF AN INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE FROM TRYPANOSOMA VIVAX IN COMPLEX WITH THE SUBSTRATE ANALOGUE 3-DEAZA-ADENOSINE | | Descriptor: | 3-DEAZA-ADENOSINE, CALCIUM ION, INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE | | Authors: | Versees, W, Decanniere, K, Pelle, R, Depoorter, J, Parkin, D.W, Steyaert, J. | | Deposit date: | 2000-12-12 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of a novel purine specific nucleoside hydrolase from Trypanosoma vivax.
J.Mol.Biol., 307, 2001
|
|
4AVZ
 
 | | Tailspike protein mutant E372Q of E. coli bacteriophage HK620 | | Descriptor: | TAIL SPIKE PROTEIN, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM | | Authors: | Gohlke, U, Broeker, N.K, Mueller, J.J, Uetrecht, C, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2012-05-30 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Single Amino Acid Exchange in Bacteriophage Hk620 Tailspike Protein Results in Thousand-Fold Increase of its Oligosaccharide Affinity.
Glycobiology, 23, 2013
|
|
4ZIA
 
 | | Crystal Structure of STAT3 N-terminal domain | | Descriptor: | FORMIC ACID, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Hu, T, Chopra, R. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Impact of the N-Terminal Domain of STAT3 in STAT3-Dependent Transcriptional Activity.
Mol.Cell.Biol., 35, 2015
|
|
1HNX
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH PACTAMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
1A2T
 
 | | STAPHYLOCOCCAL NUCLEASE, B-MERCAPTOETHANOL DISULFIDE TO V23C VARIANT | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Wynn, R, Harkins, P.C, Richards, F.M, Fox, R.O. | | Deposit date: | 1998-01-11 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Mobile unnatural amino acid side chains in the core of staphylococcal nuclease.
Protein Sci., 5, 1996
|
|
6KOS
 
 | |
4YYU
 
 | | Ficin C crystal form I | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ficin isoform C, SULFATE ION | | Authors: | Azarkan, M, Baeyens-Volant, D, Loris, R. | | Deposit date: | 2015-03-24 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.177 Å) | | Cite: | Crystal structure of four variants of the protease Ficin from the common fig
To Be Published
|
|
4YZZ
 
 | | Crystal structure of a TRAP transporter solute binding protein (IPR025997) from Bordetella bronchiseptica RB50 (BB0280, TARGET EFI-500035) mixed occupancy dimer, copurified calcium and picolinate bound active site versus apo site | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-25 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a TRAP transporter solute binding protein (IPR025997) from Bordetella bronchiseptica RB50 (BB0280, TARGET EFI-500035) mixed occupancy dimer, copurified calcium and picolinate bound active site versus apo site
To be published
|
|
4ZO3
 
 | | AidC, a Dizinc Quorum-Quenching Lactonase, in complex with a product N-hexnoyl-L-homoserine | | Descriptor: | Acylhomoserine lactonase, N-hexanoyl-L-homoserine, ZINC ION | | Authors: | Mascarenhas, R, Thomas, P.W, Wu, C.-X, Nocek, B.P, Hoang, Q, Fast, W, Liu, D. | | Deposit date: | 2015-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Biochemical Characterization of AidC, a Quorum-Quenching Lactonase with Atypical Selectivity.
Biochemistry, 54, 2015
|
|
1A3U
 
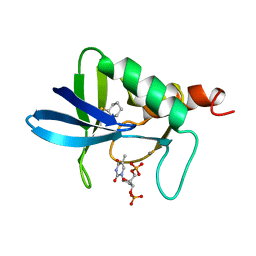 | | STAPHYLOCOCCAL NUCLEASE, CYCLOHEXANE THIOL DISULFIDE TO V23C VARIANT | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Wynn, R, Harkins, P.C, Richards, F.M, Fox, R.O. | | Deposit date: | 1998-01-24 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Comparison of straight chain and cyclic unnatural amino acids embedded in the core of staphylococcal nuclease.
Protein Sci., 6, 1997
|
|
4Z16
 
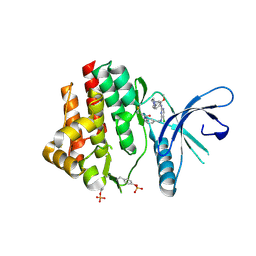 | | Crystal Structure of the Jak3 Kinase Domain Covalently Bound to N-(3-(((5-chloro-2-((2-methoxy-4-(4-methylpiperazin-1-yl)phenyl)amino)pyrimidin-4-yl)amino)methyl)phenyl)acrylamide | | Descriptor: | N-(3-{[(5-chloro-2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]methyl}phenyl)prop-2-enamide, Tyrosine-protein kinase JAK3 | | Authors: | McNally, R, Tan, L, Gray, N.S, Eck, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of Selective Covalent Janus Kinase 3 Inhibitors.
J.Med.Chem., 58, 2015
|
|
4BJH
 
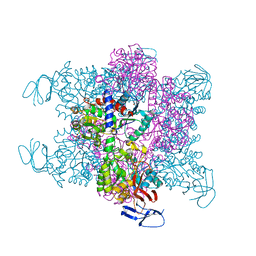 | | Crystal Structure of the Aquifex Reactor Complex Formed by Dihydroorotase (H180A, H232A) with Dihydroorotate and Aspartate Transcarbamoylase with N-(phosphonacetyl)-L-aspartate (PALA) | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, 1,2-ETHANEDIOL, ASPARTATE CARBAMOYLTRANSFERASE, ... | | Authors: | Edwards, B.F.P, Martin, P.D, Grimley, E, Vaishnav, A, Fernando, R, Brunzelle, J.S, Cordes, M, Evans, H.G, Evans, D.R. | | Deposit date: | 2013-04-18 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Mononuclear Metal Center of Type-I Dihydroorotase from Aquifex Aeolicus.
Bmc Biochem., 14, 2013
|
|
6KUK
 
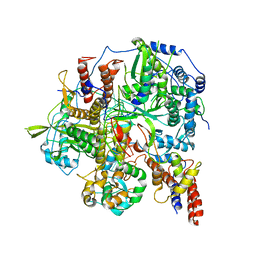 | | Structure of influenza D virus polymerase bound to vRNA promoter in mode A conformation (class A1) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
1HNZ
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH HYGROMYCIN B | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
1HYF
 
 | | RIBONUCLEASE T1 V16A MUTANT IN COMPLEX WITH SR2+ | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, GUANYL-SPECIFIC RIBONUCLEASE T1, STRONTIUM ION | | Authors: | De Swarte, J, De Vos, S, Langhorst, U, Steyaert, J, Loris, R. | | Deposit date: | 2001-01-19 | | Release date: | 2001-02-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The contribution of metal ions to the conformational stability of ribonuclease T1: crystal versus solution.
Eur.J.Biochem., 268, 2001
|
|
3N5E
 
 | | Crystal Structure of human thymidylate synthase bound to a peptide inhibitor | | Descriptor: | SULFATE ION, Synthetic peptide LR, Thymidylate synthase | | Authors: | Pozzi, C, Cardinale, D, Guaitoli, G, Tondi, D, Luciani, R, Myllykallio, H, Ferrari, S, Costi, M.P, Mangani, S. | | Deposit date: | 2010-05-25 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Protein-protein interface-binding peptides inhibit the cancer therapy target human thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1HQP
 
 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF PORCINE ODORANT-BINDING PROTEIN | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, ODORANT-BINDING PROTEIN | | Authors: | Perduca, M, Mancia, F, Del Giorgio, R, Monaco, H.L. | | Deposit date: | 2000-12-19 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a truncated form of porcine odorant-binding protein.
Proteins, 42, 2001
|
|
