3V7P
 
 | | Crystal structure of amidohydrolase nis_0429 (target efi-500396) from Nitratiruptor sp. sb155-2 | | Descriptor: | Amidohydrolase family protein, BENZOIC ACID, BICARBONATE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-21 | | Release date: | 2012-01-11 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Amidohydrolase Nis_0429 (Target Efi-500319) from Nitratiruptor Sp. Sb155-2
To be Published
|
|
4LMR
 
 | | Crystal structure of L-lactate dehydrogenase from Bacillus cereus ATCC 14579, NYSGRC Target 029452 | | Descriptor: | L-lactate dehydrogenase 1 | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-10 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of L-lactate dehydrogenase from Bacillus cereus ATCC 14579, NYSGRC Target 029452
To be Published
|
|
1WKD
 
 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-08-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile.
Biochemistry, 35, 1996
|
|
4LCA
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with thymidine | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, POTASSIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with thymidine
To be Published
|
|
2TGP
 
 | | THE GEOMETRY OF THE REACTIVE SITE AND OF THE PEPTIDE GROUPS IN TRYPSIN, TRYPSINOGEN AND ITS COMPLEXES WITH INHIBITORS | | Descriptor: | CALCIUM ION, SULFATE ION, TRYPSIN INHIBITOR, ... | | Authors: | Huber, R, Bode, W, Deisenhofer, J, Schwager, P. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors
Acta Crystallogr.,Sect.B, 39, 1983
|
|
1WKE
 
 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-08-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile.
Biochemistry, 35, 1996
|
|
4LBX
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, ADENOSINE, DIMETHYL SULFOXIDE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with cytidine
To be Published
|
|
2TPI
 
 | | ON THE DISORDERED ACTIVATION DOMAIN IN TRYPSINOGEN. CHEMICAL LABELLING AND LOW-TEMPERATURE CRYSTALLOGRAPHY | | Descriptor: | ISOLEUCINE, MERCURY (II) ION, TRYPSIN INHIBITOR, ... | | Authors: | Walter, J, Steigemann, W, Singh, T.P, Bartunik, H, Bode, W, Huber, R. | | Deposit date: | 1981-10-26 | | Release date: | 1982-03-04 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | On the Disordered Activation Domain in Trypsinogen. Chemical Labelling and Low-Temperature Crystallography
Acta Crystallogr.,Sect.B, 38, 1982
|
|
1WKF
 
 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-08-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile.
Biochemistry, 35, 1996
|
|
2UW2
 
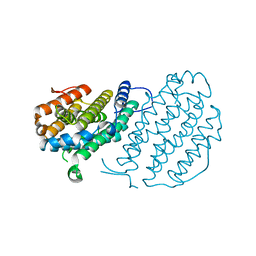 | | Crystal structure of human ribonucleotide reductase subunit R2 | | Descriptor: | FE (III) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE M2 SUBUNIT | | Authors: | Welin, M, Ogg, D, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Kotenyova, T, Magnusdottir, A, Moche, M, Nilsson-Ehle, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Stenmark, P, Uppenberg, J, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Nordlund, P. | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Ribonucleotide Reductase Subunit R2
To be Published
|
|
4M1Q
 
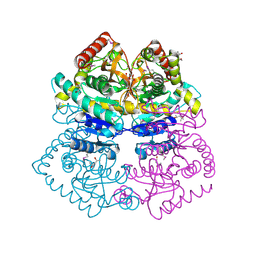 | | Crystal structure of L-lactate dehydrogenase from Bacillus selenitireducens MLS10, NYSGRC Target 029814. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, L-lactate dehydrogenase, PHOSPHATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-03 | | Release date: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of L-lactate dehydrogenase from Bacillus selenitireducens MLS10, NYSGRC Target 029814.
To be Published
|
|
4M7W
 
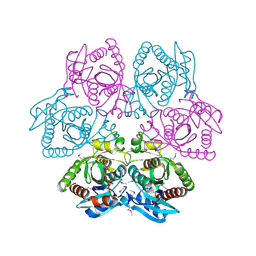 | | Crystal structure of purine nucleoside phosphorylase from Leptotrichia buccalis C-1013-b, NYSGRC Target 029767. | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-12 | | Release date: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Leptotrichia buccalis C-1013-b, NYSGRC Target 029767.
To be Published
|
|
1X9X
 
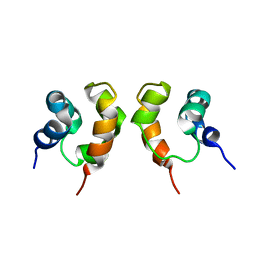 | | Solution Structure of Dimeric SAM Domain from MAPKKK Ste11 | | Descriptor: | Serine/threonine-protein kinase STE11 | | Authors: | Bhattacharjya, S, Xu, P, Gingras, R, Shaykhutdinov, R, Wu, C, Whiteway, M, Ni, F. | | Deposit date: | 2004-08-24 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the dimeric SAM domain of MAPKKK Ste11 and its interactions with the adaptor protein Ste50 from the budding yeast: implications for Ste11 activation and signal transmission through the Ste50-Ste11 complex.
J.Mol.Biol., 344, 2004
|
|
5AAM
 
 | | Structure of a redesigned cross-reactive antibody to dengue virus with increased in vivo potency | | Descriptor: | ENVELOPE PROTEIN, SCFV513 | | Authors: | Wong, Y.H, Robinson, L.N, Lescar, J, Sasisekharan, R. | | Deposit date: | 2015-07-27 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure-Guided Design of an Anti-Dengue Antibody Directed to a Non-Immunodominant Epitope.
Cell(Cambridge,Mass.), 162, 2015
|
|
6GQH
 
 | |
1XBW
 
 | | 1.9A Crystal Structure of the protein isdG from Staphylococcus aureus aureus, Structural genomics, MCSG | | Descriptor: | hypothetical protein isdG | | Authors: | Zhang, R, Wu, R, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-31 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases.
J.Biol.Chem., 280, 2005
|
|
5AAE
 
 | | Aurora A kinase bound to an imidazopyridine inhibitor (14d) | | Descriptor: | 3-((4-(6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl)-1H-pyrazol-1-yl)methyl)-5-methylisoxazole, AURORA KINASE A | | Authors: | McIntyre, P.J, Bayliss, R. | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | 7-(Pyrazol-4-Yl)-3H-Imidazo[4,5-B]Pyridine-Based Derivatives for Kinase Inhibition: Co-Crystallisation Studies with Aurora-A Reveal Distinct Differences in the Orientation of the Pyrazole N1-Substituent.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7UFV
 
 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-6766 | | Descriptor: | (3P)-N-[(1S)-3-amino-1-(3-chlorophenyl)-3-oxopropyl]-3-(2-fluorophenyl)-1H-pyrazole-4-carboxamide, DDB1- and CUL4-associated factor 1, UNKNOWN ATOM OR ION | | Authors: | Kimani, S, Li, A, Li, Y, Dong, A, Hutchinson, A, Seitova, A, Wilson, B, Al-Awar, R, Vedadi, M, Brown, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Nanomolar DCAF1 Small Molecule Ligands.
J.Med.Chem., 66, 2023
|
|
4QJY
 
 | | Crystal structure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 | | Descriptor: | ACETATE ION, GH127 beta-L-arabinofuranosidase | | Authors: | Lansky, S, Salama, R, Dann, R, Shner, I, Manjasetty, B, Belrhali, H, Shoham, Y, Shoham, G. | | Deposit date: | 2014-06-05 | | Release date: | 2015-06-10 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Crystal structure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6
To be Published
|
|
2UZ6
 
 | | AChBP-targeted a-conotoxin correlates distinct binding orientations with nAChR subtype selectivity. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-CONOTOXIN TXIA(A10L), GLYCEROL, ... | | Authors: | Ulens, C, Dutertre, S, Buttner, R, Fish, A, van Elk, R, Kendel, Y, Hopping, G, Alewood, P.F, Schroeder, C, Nicke, A, Smit, A.B, Sixma, T.K, Lewis, R.J. | | Deposit date: | 2007-04-25 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Achbp-Targeted Alpha-Conotoxin Correlates Distinct Binding Orientations with Nachr Subtype Selectivity
Embo J., 26, 2007
|
|
2VIE
 
 | | Human BACE-1 in complex with N-((1S,2R)-1-benzyl-2-hydroxy-3-((1,1,5- trimethylhexyl)amino)propyl)-3-(ethylamino)-5-(2-oxopyrrolidin-1-yl) benzamide | | Descriptor: | BETA-SECRETASE 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(1,1,5-trimethylhexyl)amino]propyl}-3-(ethylamino)-5-(2-oxopyrrolidin-1-yl)benzamide | | Authors: | Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Faller, A, Hawkins, J, Hussain, I, MacPherson, D, Maile, G, Matico, R, Milner, P, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Riddell, D, Rowland, P, Soleil, V, Smith, K, Stanway, S, Stemp, G, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Ward, J, Wayne, G. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-29 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bace-1 Inhibitors Part 2: Identification of Hydroxy Ethylamines (Heas) with Reduced Peptidic Character.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6GXK
 
 | | Crystal structure of Aldo-Keto Reductase 1C3 (AKR1C3) complexed with inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, 4-[[1-(4-chlorophenyl)carbonyl-5-methoxy-2-methyl-indol-3-yl]methyl]-1,2,5-oxadiazol-3-one, Aldo-keto reductase family 1 member C3, ... | | Authors: | Goyal, P, Wahlgren, W.Y, Friemann, R. | | Deposit date: | 2018-06-27 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bioisosteres of Indomethacin as Inhibitors of Aldo-Keto Reductase 1C3.
Acs Med.Chem.Lett., 10, 2019
|
|
7ZAU
 
 | | Fascin-1 in complex with Nb 3E11 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Fascin, ... | | Authors: | Burgess, S.G, Bayliss, R. | | Deposit date: | 2022-03-22 | | Release date: | 2023-09-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanobody inhibitor of Fascin-1 actin-bundling activity and filopodia formation.
Open Biology, 14, 2024
|
|
1NJU
 
 | | Complex structure of HCMV Protease and a peptidomimetic inhibitor | | Descriptor: | Assemblin, N-(6-aminohexanoyl)-3-methyl-L-valyl-3-methyl-L-valyl-N~1~-[(2S,3S)-3-hydroxy-4-oxo-4-{[(1R)-1-phenylpropyl]amino}butan-2-yl]-N~4~,N~4~-dimethyl-L-aspartamide | | Authors: | Khayat, R, Batra, R, Qian, C, Halmos, T, Bailey, M, Tong, L. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Studies of Inhibitor Binding to Human Cytomegalovirus Protease
Biochemistry, 42, 2003
|
|
7ZCM
 
 | |
