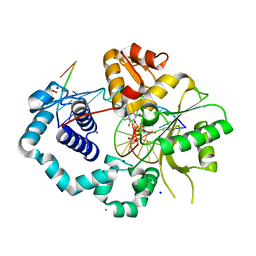
5TB9
 
 | |
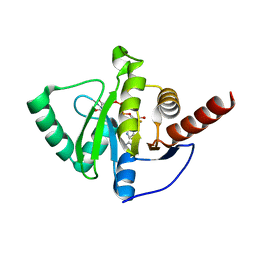
8AZL
 
 | |
5TBB
 
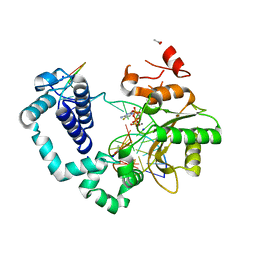 | |
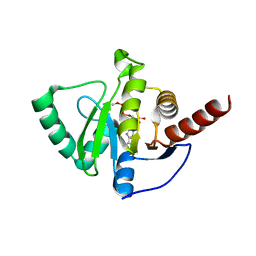
8AZP
 
 | |
8AZM
 
 | |
8AZN
 
 | |
8AXB
 
 | | Cryo-EM structure of Cas12k-sgRNA binary complex (type V-K CRISPR-associated transposon) | | 分子名称: | Cas12k, sgRNA | | 著者 | Tenjo-Castano, F, Sofos, N, Stella, S, Molina, R, Pape, T, Lopez-Mendez, B, Stutzke, L.S, Temperini, P, Montoya, G. | | 登録日 | 2022-08-31 | | 公開日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Structural prediction of the Cas12k interaction with the transposon pre-integration complex
To Be Published
|
|
1PPI
 
 | | THE ACTIVE CENTER OF A MAMMALIAN ALPHA-AMYLASE. THE STRUCTURE OF THE COMPLEX OF A PANCREATIC ALPHA-AMYLASE WITH A CARBOHYDRATE INHIBITOR REFINED TO 2.2 ANGSTROMS RESOLUTION | | 分子名称: | 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-beta-D-glucopyranose, ALPHA-AMYLASE, CALCIUM ION, ... | | 著者 | Qian, M, Haser, R, Payan, F. | | 登録日 | 1994-02-22 | | 公開日 | 1995-05-24 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The active center of a mammalian alpha-amylase. Structure of the complex of a pancreatic alpha-amylase with a carbohydrate inhibitor refined to 2.2-A resolution.
Biochemistry, 33, 1994
|
|
5TC4
 
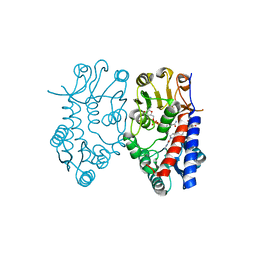 | | Crystal structure of human mitochondrial methylenetetrahydrofolate dehydrogenase-cyclohydrolase (MTHFD2) in complex with LY345899 and cofactors | | 分子名称: | 4-(7-AMINO-9-HYDROXY-1-OXO-3,3A,4,5-TETRAHYDRO-2,5,6,8,9B-PENTAAZA-CYCLOPENTA[A]NAPHTHALEN-2-YL)-PHENYLCARBONYL-GLUTAMI C ACID, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | 著者 | Gustafsson, R, Jemth, A.-S, Gustafsson Sheppard, N, Farnegardh, K, Loseva, O, Wiita, E, Bonagas, N, Dahllund, L, Llona-Minguez, S, Haggblad, M, Henriksson, M, Andersson, Y, Homan, E, Helleday, T, Stenmark, P. | | 登録日 | 2016-09-14 | | 公開日 | 2016-12-14 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Crystal Structure of the Emerging Cancer Target MTHFD2 in Complex with a Substrate-Based Inhibitor.
Cancer Res., 77, 2017
|
|
5ZCJ
 
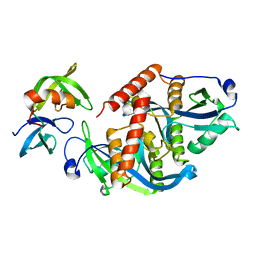 | | Crystal structure of complex | | 分子名称: | TP53-binding protein 1, Tudor-interacting repair regulator protein | | 著者 | Wang, J, Yuan, Z, Cui, Y, Xie, R, Wang, M, Ma, Y, Yu, X, Liu, X. | | 登録日 | 2018-02-17 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.004 Å) | | 主引用文献 | Crystal structure of complex
To Be Published
|
|
5TD8
 
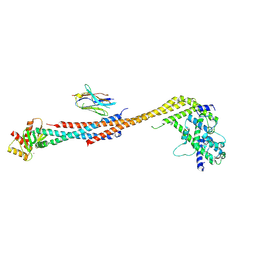 | | Crystal structure of an Extended Dwarf Ndc80 Complex | | 分子名称: | Kinetochore protein NDC80, Kinetochore protein NUF2, Kinetochore protein SPC24, ... | | 著者 | Valverde, R, Ingram, J, Harrison, S.C. | | 登録日 | 2016-09-17 | | 公開日 | 2016-11-16 | | 最終更新日 | 2019-11-20 | | 実験手法 | X-RAY DIFFRACTION (7.531 Å) | | 主引用文献 | Conserved Tetramer Junction in the Kinetochore Ndc80 Complex.
Cell Rep, 17, 2016
|
|
5YXZ
 
 | | Co-crystal Structure of KRAS (G12C) covalently bound with Quinazoline based inhibitor JBI484 | | 分子名称: | 1,2-ETHANEDIOL, 1-[4-[6-chloranyl-8-fluoranyl-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl]propan-1-one, GTPase KRas, ... | | 著者 | Swaminathan, S, Thakur, M.K, Kandan, S, Gautam, A, Kanavalli, M, Simhadri, P, Gosu, R. | | 登録日 | 2017-12-07 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Co-crystal Structure of KRAS (G12C) covalently bound with Quinazoline based inhibitor JBI484
To Be Published
|
|
5YY1
 
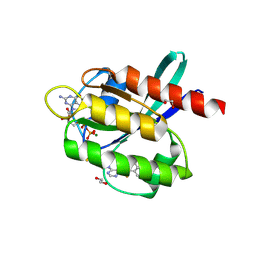 | | Co-crystal Structure of KRAS (G12C) covalently bound with Quinazoline based inhibitor JBI739 | | 分子名称: | 1,2-ETHANEDIOL, 1-[4-[6-chloranyl-8-fluoranyl-7-[2-(trifluoromethyl)phenyl]quinazolin-4-yl]piperazin-1-yl]propan-1-one, GTPase KRas, ... | | 著者 | Swaminathan, S, Thakur, M.K, Kandan, S, Gautam, A, Kanavalli, M, Simhadri, P, Gosu, R. | | 登録日 | 2017-12-07 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Co-crystal Structure of KRAS (G12C) covalently bound with Quinazoline based inhibitor JBI739
To Be Published
|
|
8B8G
 
 | | Cryo-EM structure of Ca2+-free mTMEM16F F518H mutant in Digitonin | | 分子名称: | Anoctamin-6 | | 著者 | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | 登録日 | 2022-10-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8K
 
 | | Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin closed/closed | | 分子名称: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6, CALCIUM ION | | 著者 | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | 登録日 | 2022-10-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8J
 
 | | Cryo-EM structure of Ca2+-bound mTMEM16F F518H mutant in Digitonin | | 分子名称: | Anoctamin-6, CALCIUM ION | | 著者 | Arndt, M, Alvadia, C, Straub, M.S, Clerico Mosina, V, Paulino, C, Dutzler, R. | | 登録日 | 2022-10-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (2.96 Å) | | 主引用文献 | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8BC0
 
 | | Cryo-EM structure of Ca2+-bound mTMEM16F F518A Q623A mutant in GDN open/closed | | 分子名称: | Anoctamin-6, CALCIUM ION | | 著者 | Arndt, M, Alvadia, C, Straub, M, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | 登録日 | 2022-10-14 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.09 Å) | | 主引用文献 | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8BAY
 
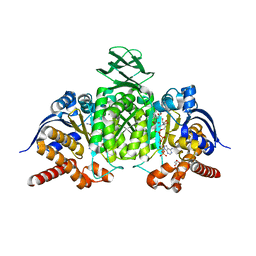 | | Crystal Structure of IDH1 variant R132C S280F in complex with NADPH, Ca2+ and 3-butyl-2-oxoglutarate | | 分子名称: | (R)-3-butyl-2-oxopentanedioic acid, (S)-3-butyl-2-oxopentanedioic acid, CALCIUM ION, ... | | 著者 | Rabe, P, Schofield, C.J, Reinbold, R, Brewitz, L. | | 登録日 | 2022-10-12 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Natural and synthetic 2-oxoglutarate derivatives are substrates for oncogenic variants of human isocitrate dehydrogenase 1 and 2.
J.Biol.Chem., 299, 2023
|
|
5YZB
 
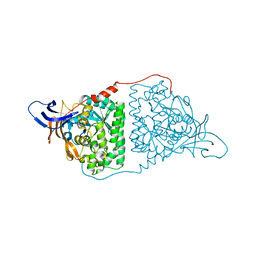 | | Crystal Structure of Human CRMP-2 with S522D-T509D-T514D-S518D mutations crystallized with GSK3b | | 分子名称: | Dihydropyrimidinase-related protein 2 | | 著者 | Imasaki, T, Sumi, T, Aoki, M, Sakai, N, Nitta, E, Shirouzu, M, Nitta, R. | | 登録日 | 2017-12-13 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Insights into the Altering Function of CRMP2 by Phosphorylation.
Cell Struct. Funct., 43, 2018
|
|
5YWY
 
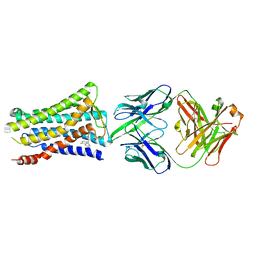 | | Crystal structure of the human prostaglandin E receptor EP4 in complex with Fab and ONO-AE3-208 | | 分子名称: | 4-[4-cyano-2-[[(2R)-2-(4-fluoranylnaphthalen-1-yl)propanoyl]amino]phenyl]butanoic acid, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | 著者 | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | 登録日 | 2017-11-30 | | 公開日 | 2018-12-05 | | 最終更新日 | 2018-12-19 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
8BC1
 
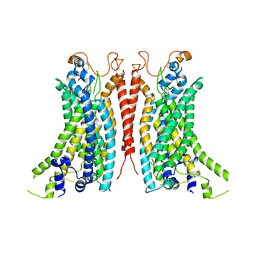 | | Cryo-EM Structure of Ca2+-bound mTMEM16F F518A_Q623A mutant in GDN | | 分子名称: | Anoctamin-6,mTMEM16F, CALCIUM ION | | 著者 | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | 登録日 | 2022-10-14 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
1NY7
 
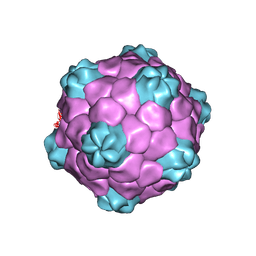 | | COWPEA MOSAIC VIRUS (CPMV) | | 分子名称: | COWPEA MOSAIC VIRUS, LARGE (L) SUBUNIT, SMALL (S) SUBUNIT | | 著者 | Lin, T, Chen, Z, Usha, R, Stauffacher, C.V, Dai, J.-B, Schmidt, T, Johnson, J.E. | | 登録日 | 2003-02-11 | | 公開日 | 2003-03-18 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The Refined Crystal Structure of Cowpea Mosaic Virus at 2.8A Resolution
Virology, 265, 1999
|
|
8B8M
 
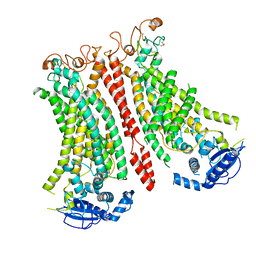 | | Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin open/closed | | 分子名称: | Anoctamin-6, CALCIUM ION | | 著者 | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | 登録日 | 2022-10-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8BK0
 
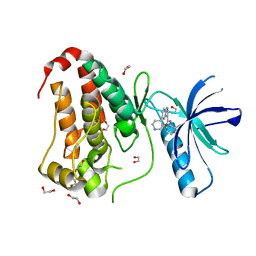 | | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with LDN-211904 | | 分子名称: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, ~{N}-(2-chlorophenyl)-6-piperidin-4-yl-imidazo[1,2-a]pyridine-3-carboxamide | | 著者 | Zhubi, R, Gerninghaus, J, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | 登録日 | 2022-11-08 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with LDN-211904
To Be Published
|
|
5TAW
 
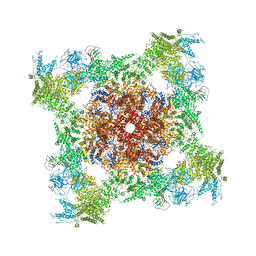 | | Structure of rabbit RyR1 (ryanodine dataset, all particles) | | 分子名称: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | 著者 | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | 登録日 | 2016-09-10 | | 公開日 | 2016-10-12 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
