4ECJ
 
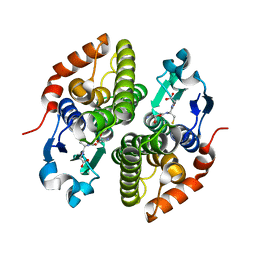 | | Crystal structure of glutathione s-transferase prk13972 (target efi-501853) from pseudomonas aeruginosa pacs2 complexed with glutathione | | Descriptor: | GLUTATHIONE, glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase Prk13972 from Pseudomonas Aeruginosa
To be Published
|
|
1CP8
 
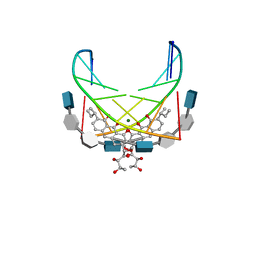 | | NMR STRUCTURE OF DNA (5'-D(TTGGCCAA)2-3') COMPLEXED WITH NOVEL ANTITUMOR DRUG UCH9 | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYDROXY-7-(SEC-BUTYL)-ANTHRACENE, DNA (5'-D(P*TP*TP*GP*GP*CP*CP*AP*A)-3'), MAGNESIUM ION, ... | | Authors: | Katahira, R, Katahira, M, Yamashita, Y, Ogawa, H, Kyogoku, Y, Yoshida, M. | | Deposit date: | 1999-06-11 | | Release date: | 1999-07-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the novel antitumor drug UCH9 complexed with d(TTGGCCAA)2 as determined by NMR.
Nucleic Acids Res., 26, 1998
|
|
6YVW
 
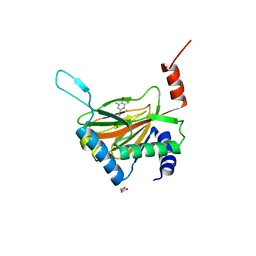 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with monocyclic BB-328 | | Descriptor: | 4-[(5-bromanyl-4,6-dimethyl-pyridin-2-yl)amino]-4-oxidanylidene-butanoic acid, Egl nine homolog 1, FE (III) ION, ... | | Authors: | Chowdhury, R, Banerji, B, Schofield, C.J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Use of cyclic peptides to induce crystallization: case study with prolyl hydroxylase domain 2.
Sci Rep, 10, 2020
|
|
1CJS
 
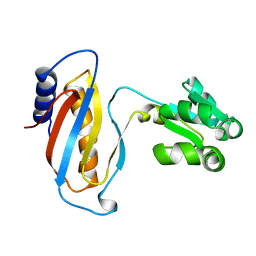 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | 50S RIBOSOMAL PROTEIN L1P | | Authors: | Nevskaya, N, Tishchenko, S, Fedorov, R, Al-Karadaghi, S, Liljas, A, Kraft, A, Piendl, W, Garber, M, Nikonov, S. | | Deposit date: | 1999-04-19 | | Release date: | 2000-05-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Archaeal ribosomal protein L1: the structure provides new insights into RNA binding of the L1 protein family.
Structure Fold.Des., 8, 2000
|
|
6YW1
 
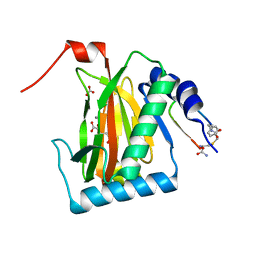 | |
1CQX
 
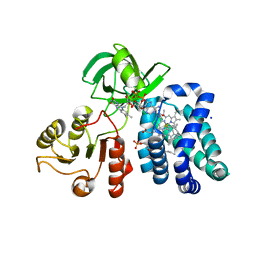 | | Crystal structure of the flavohemoglobin from Alcaligenes eutrophus at 1.75 A resolution | | Descriptor: | 1-[GLYCEROLYLPHOSPHONYL]-2-[8-(2-HEXYL-CYCLOPROPYL)-OCTANAL-1-YL]-3-[HEXADECANAL-1-YL]-GLYCEROL, FLAVIN-ADENINE DINUCLEOTIDE, FLAVOHEMOPROTEIN, ... | | Authors: | Ermler, U, Siddiqui, R.A, Cramm, R, Friedrich, B. | | Deposit date: | 1999-08-12 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the flavohemoglobin from Alcaligenes eutrophus at 1.75 A resolution.
EMBO J., 14, 1995
|
|
1CM0
 
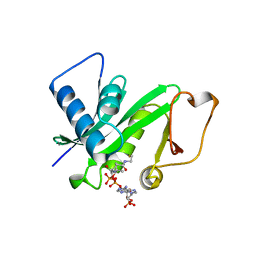 | | CRYSTAL STRUCTURE OF THE PCAF/COENZYME-A COMPLEX | | Descriptor: | COENZYME A, P300/CBP ASSOCIATING FACTOR | | Authors: | Clements, A, Rojas, J.R, Trievel, R.C, Wang, L, Berger, S.L, Marmorstein, R. | | Deposit date: | 1999-05-12 | | Release date: | 1999-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A.
EMBO J., 18, 1999
|
|
3IHK
 
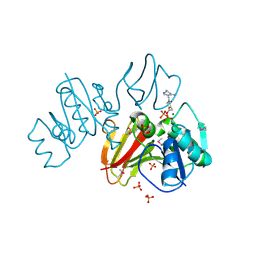 | | Crystal Structure of thiamin pyrophosphokinase from S.mutans, Northeast Structural Genomics Consortium Target SmR83 | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Kuzin, A, Abashidze, M, Seetharaman, J, Vorobiev, S, Mao, M, Xiao, R, Ciccosanti, C, Maglaqui, M, Foote, E.L, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-25 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Northeast Structural Genomics Consortium Target SmR83
To be published
|
|
4MF5
 
 | | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with traces of one GSH bound | | Descriptor: | FORMIC ACID, GLUTATHIONE, Glutathione S-transferase domain | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with traces of one GSH bound
To be Published
|
|
3VOS
 
 | | Crystal structure of Aspartate semialdehyde dehydrogenase Complexed With glycerol and sulfate From Mycobacterium tuberculosis H37Rv | | Descriptor: | Aspartate-semialdehyde dehydrogenase, GLYCEROL, SULFATE ION | | Authors: | Vyas, R, Tewari, R, Weiss, M.S, Karthikeyan, S. | | Deposit date: | 2012-02-07 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of ternary complexes of aspartate-semialdehyde dehydrogenase (Rv3708c) from Mycobacterium tuberculosis H37Rv
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6KUT
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B2) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
1CS3
 
 | |
6KXN
 
 | | Crystal structure of W50A mutant of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ueda, M, Shimosaka, M, Arai, R. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of CsChiL, a chitinase from Chitiniphilus shinanonensis
To be published
|
|
3VX6
 
 | | Crystal structure of Kluyveromyces marxianus Atg7NTD | | Descriptor: | E1 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1CWX
 
 | | SOLUTION STRUCTURE OF THE HEPATITIS C VIRUS N-TERMINAL CAPSID PROTEIN 2-45 [C-HCV(2-45)] | | Descriptor: | HEPATITIS C VIRUS CAPSID PROTEIN | | Authors: | Ladaviere, L, Deleage, G, Montserret, R, Dalbon, P, Jolivet, M, Penin, F. | | Deposit date: | 1999-08-27 | | Release date: | 1999-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Immunodominant Antigenic Region of the Hepatitis C Virus Capsid Protein by NMR
To be Published
|
|
3V7P
 
 | | Crystal structure of amidohydrolase nis_0429 (target efi-500396) from Nitratiruptor sp. sb155-2 | | Descriptor: | Amidohydrolase family protein, BENZOIC ACID, BICARBONATE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-21 | | Release date: | 2012-01-11 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Amidohydrolase Nis_0429 (Target Efi-500319) from Nitratiruptor Sp. Sb155-2
To be Published
|
|
1CX8
 
 | | CRYSTAL STRUCTURE OF THE ECTODOMAIN OF HUMAN TRANSFERRIN RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SAMARIUM (III) ION, TRANSFERRIN RECEPTOR PROTEIN | | Authors: | Lawrence, C.M, Ray, S, Babyonyshev, M, Galluser, R, Borhani, D, Harrison, S.C. | | Deposit date: | 1999-08-28 | | Release date: | 1999-09-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the ectodomain of human transferrin receptor.
Science, 286, 1999
|
|
3OBH
 
 | | X-ray crystal structure of protein SP_0782 (7-79) from Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR104 | | Descriptor: | ACETIC ACID, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuzin, A, Abashidze, M, Lew, S, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-08-06 | | Release date: | 2010-09-15 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | X-ray crystal structure of protein SP_0782 (7-79) from Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR104
To be Published
|
|
2WW5
 
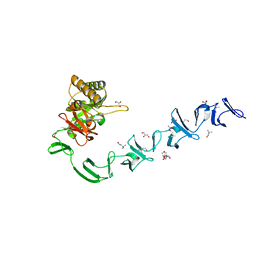 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae at 1.6 A resolution | | Descriptor: | 1,4-BETA-N-ACETYLMURAMIDASE, CHLORIDE ION, CHOLINE ION, ... | | Authors: | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L, Menendez, M. | | Deposit date: | 2009-10-21 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1D2I
 
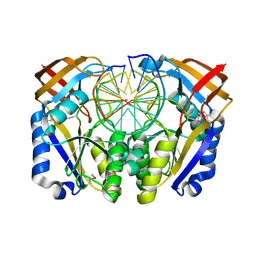 | | CRYSTAL STRUCTURE OF RESTRICTION ENDONUCLEASE BGLII COMPLEXED WITH DNA 16-MER | | Descriptor: | DNA (5'-D(*TP*AP*TP*TP*AP*TP*AP*GP*AP*TP*CP*TP*AP*TP*AP*A)-3'), MAGNESIUM ION, PROTEIN (RESTRICTION ENDONUCLEASE BGLII) | | Authors: | Lukacs, C.M, Kucera, R, Schildkraut, I, Aggarwal, A.K. | | Deposit date: | 1999-09-23 | | Release date: | 2000-02-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 A resolution.
Nat.Struct.Biol., 7, 2000
|
|
3I7J
 
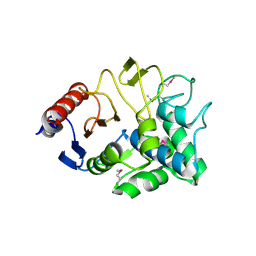 | | Crystal Structure of a beta-lactamase (Mb2281c) from Mycobacterium bovis, Northeast Structural Genomics Consortium Target MbR246 | | Descriptor: | beta-lactamase Mb2281c | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-08 | | Release date: | 2009-07-14 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Northeast Structural Genomics Consortium Target MbR246
To be Published
|
|
6YW2
 
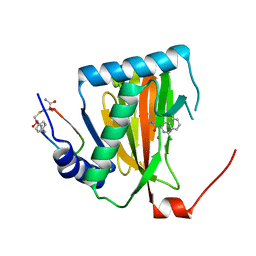 | |
3NNG
 
 | | Crystal structure of the F5/8 type C domain of Q5LFR2_BACFN protein from Bacteroides fragilis. Northeast Structural Genomics Consortium Target BfR258E | | Descriptor: | CALCIUM ION, uncharacterized protein | | Authors: | Vorobiev, S, Su, M, Dimaio, F, Baker, D, Seetharaman, J, Janjua, J, Xiao, R, Ciccosanti, C, Foote, E.L, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-23 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Crystal structure of the F5/8 type C domain of Q5LFR2_BACFN protein from Bacteroides fragilis.
To be Published
|
|
4Q49
 
 | |
6YW3
 
 | |
