1THR
 
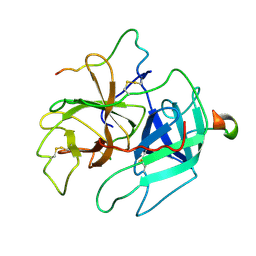 | | STRUCTURES OF THROMBIN COMPLEXES WITH A DESIGNED AND A NATURAL EXOSITE INHIBITOR | | Descriptor: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), HIRULLIN | | Authors: | Qiu, X, Yin, M, Padmanabhan, K.P, Krstenansky, J.L, Tulinsky, A. | | Deposit date: | 1993-06-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of thrombin complexes with a designed and a natural exosite peptide inhibitor.
J.Biol.Chem., 268, 1993
|
|
6JH7
 
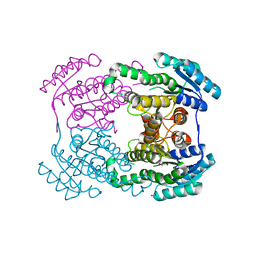 | | Crystal structure of AerF from Microcystis aeruginosa | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Short chain dehydrogenase family protein, ... | | Authors: | Qiu, X. | | Deposit date: | 2019-02-17 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural and functional investigation of AerF, a NADPH-dependent alkenal double bond reductase participating in the biosynthesis of Choi moiety of aeruginosin
J.Struct.Biol., 2019
|
|
6JHB
 
 | |
6JHA
 
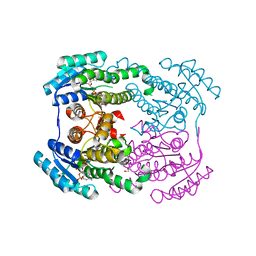 | | Crystal structure of NADPH bound AerF from Microcystis aeruginosa | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase family protein, TRIETHYLENE GLYCOL | | Authors: | Qiu, X. | | Deposit date: | 2019-02-17 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and functional investigation of AerF, a NADPH-dependent alkenal double bond reductase participating in the biosynthesis of Choi moiety of aeruginosin
J.Struct.Biol., 2019
|
|
1JIL
 
 | | Crystal structure of S. aureus TyrRS in complex with SB284485 | | Descriptor: | [2-AMINO-3-(4-HYDROXY-PHENYL)-PROPIONYLAMINO]- (3,4,5-TRIHYDROXY-6-METHYL-TETRAHYDRO-PYRAN-2-YL)- ACETIC ACID, tyrosyl-tRNA synthetase | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Jarvest, R.L. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors.
Protein Sci., 10, 2001
|
|
1ABI
 
 | |
1JIJ
 
 | | Crystal structure of S. aureus TyrRS in complex with SB-239629 | | Descriptor: | [2-AMINO-3-(4-HYDROXY-PHENYL)-PROPIONYLAMINO]-(1,3,4,5-TETRAHYDROXY-4-HYDROXYMETHYL-PIPERIDIN-2-YL)- ACETIC ACID, tyrosyl-tRNA synthetase | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Jarvest, R.L. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors.
Protein Sci., 10, 2001
|
|
1JII
 
 | | Crystal structure of S. aureus TyrRS in complex with SB-219383 | | Descriptor: | [2-AMINO-3-(4-HYDROXY-PHENYL)-PROPIONYLAMINO]- (2,4,5,8-TETRAHYDROXY-7-OXA-2-AZA-BICYCLO[3.2.1]OCT-3-YL)- ACETIC ACID, tyrosyl-tRNA synthetase | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Jarvest, R.L. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors.
Protein Sci., 10, 2001
|
|
1ABJ
 
 | |
1JIK
 
 | | Crystal structure of S. aureus TyrRS in complex with SB-243545 | | Descriptor: | [2-AMINO-3-(4-HYDROXY-PHENYL)-PROPIONYLAMINO]-(1,3,4,5-TETRAHYDROXY-4-HYDROXYMETHYL-PIPERIDIN-2-YL)- ACETIC ACID BUTYL ESTER, tyrosyl-tRNA synthetase | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Jarvest, R.L. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors.
Protein Sci., 10, 2001
|
|
2OBD
 
 | | Crystal Structure of Cholesteryl Ester Transfer Protein | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Qiu, X. | | Deposit date: | 2006-12-18 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1T8K
 
 | | Crystal Structure of apo acyl carrier protein from E. coli | | Descriptor: | Acyl carrier protein, IMIDAZOLE, ZINC ION | | Authors: | Qiu, X, Janson, C.A. | | Deposit date: | 2004-05-13 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of apo acyl carrier protein and a proposal to engineer protein crystallization through metal ions.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TMB
 
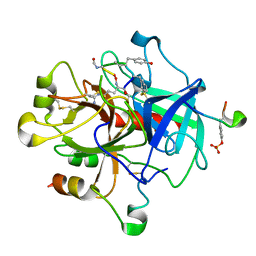 | | MOLECULAR BASIS FOR THE INHIBITION OF HUMAN ALPHA-THROMBIN BY THE MACROCYCLIC PEPTIDE CYCLOTHEONAMIDE A | | Descriptor: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), HIRUGEN, ... | | Authors: | Qiu, X, Padmanabhan, K.P, Maryanoff, B.E, Tulinsky, A. | | Deposit date: | 1993-05-27 | | Release date: | 1994-01-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the inhibition of human alpha-thrombin by the macrocyclic peptide cyclotheonamide A.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1ZOW
 
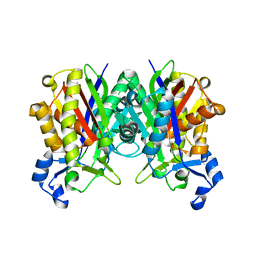 | | Crystal Structure of S. aureus FabH, beta-ketoacyl carrier protein synthase III | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase III | | Authors: | Qiu, X, Choudhry, A.E, Janson, C.A, Grooms, M, Daines, R.A, Lonsdale, J.T, Khandekar, S.S. | | Deposit date: | 2005-05-15 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and substrate specificity of the beta-ketoacyl-acyl carrier protein synthase III (FabH) from Staphylococcus aureus.
Protein Sci., 14, 2005
|
|
5ID6
 
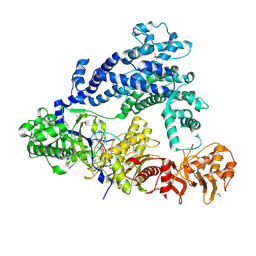 | | Structure of Cpf1/RNA Complex | | Descriptor: | Cpf1, MAGNESIUM ION, RNA (5'-R(P*AP*AP*UP*UP*UP*CP*UP*AP*CP*UP*AP*AP*GP*UP*GP*UP*AP*GP*AP*UP*C)-3') | | Authors: | Dong, D, Ren, K, Qiu, X, Wang, J, Huang, Z. | | Deposit date: | 2016-02-24 | | Release date: | 2016-04-27 | | Last modified: | 2016-05-11 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | The crystal structure of Cpf1 in complex with CRISPR RNA
Nature, 532, 2016
|
|
5GZO
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
5GZN
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody Heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
7KDP
 
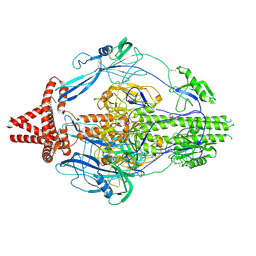 | | HCMV prefusion gB in complex with fusion inhibitor WAY-174865 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein B, ... | | Authors: | Liu, Y, Heim, P.K, Che, Y, Chi, X, Qiu, X, Han, S, Dormitzer, P.R, Yang, X. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Prefusion structure of human cytomegalovirus glycoprotein B and structural basis for membrane fusion.
Sci Adv, 7, 2021
|
|
7KDD
 
 | | HCMV postfusion gB in complex with SM5-1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein B, ... | | Authors: | Liu, Y, Heim, P.K, Che, Y, Chi, X, Qiu, X, Han, S, Dormitzer, P.R, Yang, X. | | Deposit date: | 2020-10-08 | | Release date: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Prefusion structure of human cytomegalovirus glycoprotein B and structural basis for membrane fusion.
Sci Adv, 7, 2021
|
|
4EWS
 
 | |
5BTR
 
 | | Crystal structure of SIRT1 in complex with resveratrol and an AMC-containing peptide | | Descriptor: | AMC-containing peptide, NAD-dependent protein deacetylase sirtuin-1, RESVERATROL, ... | | Authors: | Cao, D, Wang, M, Qiu, X, Liu, D, Jiang, H, Yang, N, Xu, R.M. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for allosteric, substrate-dependent stimulation of SIRT1 activity by resveratrol
Genes Dev., 29, 2015
|
|
1LX6
 
 | |
8IVQ
 
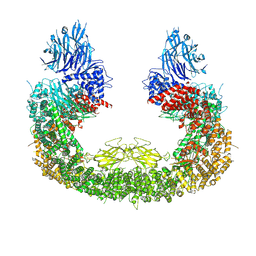 | | Cryo-EM structure of mouse BIRC6, Global map | | Descriptor: | Isoform 2 of Baculoviral IAP repeat-containing protein 6 | | Authors: | Liu, S, Jiang, T, Bu, F, Zhao, J, Wang, G, Li, N, Gao, N, Qiu, X. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanisms underlying the BIRC6-mediated regulation of apoptosis and autophagy.
Nat Commun, 15, 2024
|
|
1AT3
 
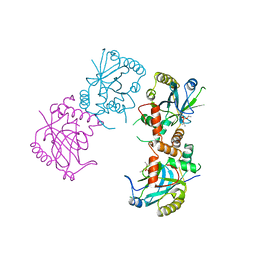 | | HERPES SIMPLEX VIRUS TYPE II PROTEASE | | Descriptor: | DIISOPROPYL PHOSPHONATE, HERPES SIMPLEX VIRUS TYPE II PROTEASE | | Authors: | Hoog, S, Smith, W.W, Qiu, X, Abdel-Meguid, S.S. | | Deposit date: | 1997-08-16 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active site cavity of herpesvirus proteases revealed by the crystal structure of herpes simplex virus protease/inhibitor complex.
Biochemistry, 36, 1997
|
|
1A36
 
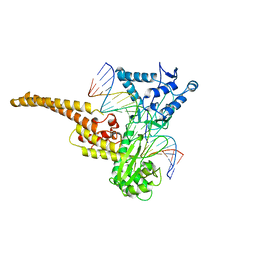 | | TOPOISOMERASE I/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*AP*GP*AP*AP*AP*AP*A P*TP*TP*TP*TP*T)- 3'), DNA (5'-D(*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*TP*AP*AP*GP*TP*C P*TP*TP*TP*TP*T)- 3'), TOPOISOMERASE I | | Authors: | Stewart, L, Redinbo, M.R, Qiu, X, Champoux, J.J, Hol, W.G.J. | | Deposit date: | 1998-01-29 | | Release date: | 1998-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A model for the mechanism of human topoisomerase I.
Science, 279, 1998
|
|
