3K1A
 
 | | Insights into substrate binding at FeMo-cofactor in nitrogenase from the structure of an alpha-70Ile MoFe protein variant | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(7)-MO-S(9)-N CLUSTER, ... | | Authors: | Peters, J.W, Sarma, R, Barney, B.M, Keable, S, Seefeldt, L.C, Dean, D.R. | | Deposit date: | 2009-09-26 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Insights into substrate binding at FeMo-cofactor in nitrogenase from the structure of an alpha-70(Ile) MoFe protein variant
J.Inorg.Biochem., 104, 2010
|
|
2MIN
 
 | | NITROGENASE MOFE PROTEIN FROM AZOTOBACTER VINELANDII, OXIDIZED STATE | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Peters, J.W, Stowell, M.H.B, Soltis, S.M, Day, M.W, Kim, J, Rees, D.C. | | Deposit date: | 1996-12-20 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Redox-dependent structural changes in the nitrogenase P-cluster.
Biochemistry, 36, 1997
|
|
3MIN
 
 | | NITROGENASE MOFE PROTEIN FROM AZOTOBACTER VINELANDII, OXIDIZED STATE | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Peters, J.W, Stowell, M.H.B, Soltis, S.M, Day, M.W, Kim, J, Rees, D.C. | | Deposit date: | 1996-12-20 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Redox-dependent structural changes in the nitrogenase P-cluster.
Biochemistry, 36, 1997
|
|
1FEH
 
 | | FE-ONLY HYDROGENASE FROM CLOSTRIDIUM PASTEURIANUM | | Descriptor: | 2 IRON/2 SULFUR/5 CARBONYL/2 WATER INORGANIC CLUSTER, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Peters, J.W, Lanzilotta, W.N, Lemon, B.J, Seefeldt, L.C. | | Deposit date: | 1998-10-28 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution.
Science, 282, 1998
|
|
1LRV
 
 | |
7SEZ
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with m7GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DNA repair NTP-phosphohydrolase, SODIUM ION | | Authors: | Peters, J.K, Tibble, R.W, Warminski, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.70001245 Å) | | Cite: | Structure of the poxvirus decapping enzyme D9 reveals its mechanism of cap recognition and catalysis.
Structure, 30, 2022
|
|
7T7H
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with inhibitor CP100356 | | Descriptor: | 4-(6,7-dimethoxy-3,4-dihydroisoquinolin-2(1H)-yl)-N-[2-(3,4-dimethoxyphenyl)ethyl]-6,7-dimethoxyquinazolin-2-amine, DNA repair NTP-phosphohydrolase, SODIUM ION | | Authors: | Peters, J.K, Gross, J.D. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78000259 Å) | | Cite: | Fluorescence-Based Activity Screening Assay Reveals Small Molecule Inhibitors of Vaccinia Virus mRNA Decapping Enzyme D9.
Acs Chem.Biol., 17, 2022
|
|
7SF0
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with trinucleotide substrate | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DNA repair NTP-phosphohydrolase, MAGNESIUM ION, ... | | Authors: | Peters, J.K, Tibble, R.W, Warminski, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95000446 Å) | | Cite: | Structure of the poxvirus decapping enzyme D9 reveals its mechanism of cap recognition and catalysis.
Structure, 30, 2022
|
|
7OIL
 
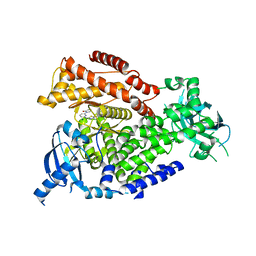 | | mPI3Kd in complex with compound 58 | | Descriptor: | 2-[(1S)-1-cyclopropylethyl]-5-[4-methyl-2-[[6-(2-oxidanylidenepyrrolidin-1-yl)pyridin-2-yl]amino]-1,3-thiazol-5-yl]-7-methylsulfonyl-3H-isoindol-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, SODIUM ION | | Authors: | Petersen, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of AZD8154, a Dual PI3K gamma delta Inhibitor for the Treatment of Asthma.
J.Med.Chem., 64, 2021
|
|
7OI4
 
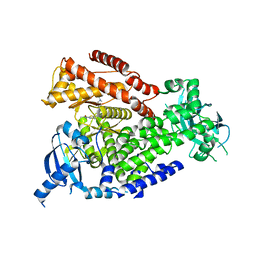 | | mPI3Kd in complex with compound 12 | | Descriptor: | N-[5-[2-[(1S)-1-cyclopropylethyl]-7-[[4-[(dimethylamino)methyl]phenyl]sulfamoyl]-1-oxidanylidene-3H-isoindol-5-yl]-4-methyl-1,3-thiazol-2-yl]ethanamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of AZD8154, a Dual PI3K gamma delta Inhibitor for the Treatment of Asthma.
J.Med.Chem., 64, 2021
|
|
7OIS
 
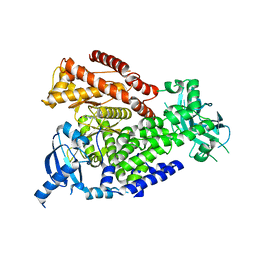 | | mPI3Kd in complex with compound 7 | | Descriptor: | N-[5-[2-[(1S)-1-cyclopropylethyl]-7-[(3-methylsulfonylphenyl)sulfamoyl]-1-oxidanylidene-3H-isoindol-5-yl]-4-methyl-1,3-thiazol-2-yl]ethanamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, SODIUM ION | | Authors: | Petersen, J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of AZD8154, a Dual PI3K gamma delta Inhibitor for the Treatment of Asthma.
J.Med.Chem., 64, 2021
|
|
7OIJ
 
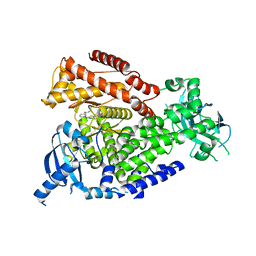 | | mPI3Kd in complex with an inhibitor | | Descriptor: | 6-[[5-[2-[(1S)-1-cyclopropylethyl]-7-(methylsulfamoyl)-1-oxidanylidene-3H-isoindol-5-yl]-4-methyl-1,3-thiazol-2-yl]amino]-N-[3-(dimethylamino)propyl]pyridine-2-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, SODIUM ION, ... | | Authors: | Petersen, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of AZD8154, a Dual PI3K gamma delta Inhibitor for the Treatment of Asthma.
J.Med.Chem., 64, 2021
|
|
2XYA
 
 | | Non-covalent inhibtors of rhinovirus 3C protease. | | Descriptor: | 2-PHENYLQUINOLIN-4-OL, PICORNAIN 3C | | Authors: | Petersen, J, Edman, K, Edfeldt, F, Johansson, C. | | Deposit date: | 2010-11-16 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Non-Covalent Inhibitors of Rhinovirus 3C Protease.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6FH5
 
 | | PI3Kg IN COMPLEX WITH Compound 7 | | Descriptor: | 3-methyl-1-(oxan-4-yl)-8-pyridin-3-yl-imidazo[4,5-c]quinolin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Petersen, J, Barlaam, B. | | Deposit date: | 2018-01-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery and Optimisation of a Novel Series of 3-Cinnoline Carboxamides as Orally Bioavailable, Highly Potent and Selective Ataxia Telangiectasia Mutated (ATM) inhibitors
To Be Published
|
|
6FTN
 
 | | mPI3Kd IN COMPLEX WITH AZ2 | | Descriptor: | Phosphor inositol 3 kinase, ~{N}-[5-[2-[(1~{S})-1-cyclopropylethyl]-7-methyl-1-oxidanylidene-3~{H}-isoindol-5-yl]-4-methyl-1,3-thiazol-2-yl]ethanamide | | Authors: | Petersen, J. | | Deposit date: | 2018-02-22 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Highly Isoform Selective Orally Bioavailable Phosphoinositide 3-Kinase (PI3K)-gamma Inhibitors.
J. Med. Chem., 61, 2018
|
|
9BF9
 
 | | Human LAG-3-HLA-DR1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, HLA class II histocompatibility antigen DR beta chain, ... | | Authors: | Petersen, J, Rossjohn, J. | | Deposit date: | 2024-04-17 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the human LAG-3-HLA-DR1-peptide complex.
Sci Immunol, 9, 2024
|
|
6GY0
 
 | | mPI3Kd IN COMPLEX WITH AZ3 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[4-methyl-5-(1-oxidanylidene-7-sulfamoyl-isoindol-5-yl)-1,3-thiazol-2-yl]ethanamide | | Authors: | Petersen, J. | | Deposit date: | 2018-06-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A class of highly selective inhibitors bind to an active state of PI3K gamma.
Nat.Chem.Biol., 15, 2019
|
|
9FRV
 
 | | Arginase 2 in complex with inhibitor | | Descriptor: | Arginase-2, mitochondrial, MANGANESE (II) ION, ... | | Authors: | Petersen, j. | | Deposit date: | 2024-06-19 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Design and Synthesis of Acyclic Boronic Acid Arginase Inhibitors.
J.Med.Chem., 67, 2024
|
|
6GQ7
 
 | | PI3Kg IN COMPLEX WITH INH | | Descriptor: | 3-methyl-1-(oxan-4-yl)-8-pyridin-3-yl-imidazo[4,5-c]quinolin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Petersen, J. | | Deposit date: | 2018-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | PI3Kg IN COMPLEX WITH INH
To Be Published
|
|
7QB1
 
 | | PPARg in complex with inhibitor | | Descriptor: | 3-[(3,4-dichlorophenyl)methyl]-4-oxidanylidene-6-phenoxy-phthalazine-1-carboxylic acid, Peroxisome proliferator-activated receptor gamma, SULFATE ION | | Authors: | Petersen, J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-05-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery by Virtual Screening of an Inhibitor of CDK5-Mediated PPAR gamma Phosphorylation.
Acs Med.Chem.Lett., 13, 2022
|
|
6XP6
 
 | | 3C11-DQ2-glia-a2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3.C11 IgH Fab, ... | | Authors: | Petersen, J, Rossjohn, J. | | Deposit date: | 2020-07-08 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A high-affinity human TCR-like antibody detects celiac disease gluten peptide-MHC complexes and inhibits T cell activation.
Sci Immunol, 6, 2021
|
|
8RFA
 
 | | Arginase 2 in complex with an inhibitor | | Descriptor: | Arginase-2, mitochondrial, MANGANESE (II) ION, ... | | Authors: | Petersen, J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of (2 R ,4 R )-4-(( S )-2-Amino-3-methylbutanamido)-2-(4-boronobutyl)pyrrolidine-2-carboxylic Acid (AZD0011), an Actively Transported Prodrug of a Potent Arginase Inhibitor to Treat Cancer.
J.Med.Chem., 67, 2024
|
|
8QFZ
 
 | | TSLP-Bicycle complex | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, CYS-HIS-TRP-LEU-GLU-ASN-CYS-TRP-ARG-GLY-PHE-CYS, Thymic stromal lymphopoietin | | Authors: | Petersen, J. | | Deposit date: | 2023-09-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Characterization of a Bicyclic Peptide (Bicycle) Binder to Thymic Stromal Lymphopoietin.
J.Med.Chem., 67, 2024
|
|
6MFF
 
 | | HLA-DQ2-glia-omega1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DQ alpha 1 chain, ... | | Authors: | Petersen, J, Ciacchi, L, Rossjohn, J. | | Deposit date: | 2018-09-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discriminative T-cell receptor recognition of highly homologous HLA-DQ2-bound gluten epitopes.
J. Biol. Chem., 294, 2019
|
|
6MFG
 
 | | HLA-DQ2-glia-alpha1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DQ alpha 1 chain, ... | | Authors: | Petersen, J, Ciacchi, L, Rossjohn, J. | | Deposit date: | 2018-09-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discriminative T-cell receptor recognition of highly homologous HLA-DQ2-bound gluten epitopes.
J. Biol. Chem., 294, 2019
|
|
