1HWV
 
 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1S)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-10 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
1HX4
 
 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1R)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-11 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
6UCD
 
 | |
5J9G
 
 | |
4IUR
 
 | |
5KRS
 
 | | HIV-1 Integrase Catalytic Core Domain in Complex with an Allosteric Inhibitor, 3-(1H-pyrrol-1-yl)-2-thiophenecarboxylic acid | | Descriptor: | 3-pyrrol-1-ylthiophene-2-carboxylic acid, DIMETHYL SULFOXIDE, Integrase | | Authors: | Patel, D, Bauman, J.D, Arnold, E. | | Deposit date: | 2016-07-07 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A New Class of Allosteric HIV-1 Integrase Inhibitors Identified by Crystallographic Fragment Screening of the Catalytic Core Domain.
J.Biol.Chem., 291, 2016
|
|
5KRT
 
 | |
134D
 
 | |
139D
 
 | |
5FII
 
 | | Structure of a human aspartate kinase, chorismate mutase and TyrA domain. | | Descriptor: | PHENYLALANINE, PHENYLALANINE-4-HYDROXYLASE | | Authors: | Patel, D, Kopec, J, Shrestha, L, Fitzpatrick, F, Pinkas, D, Chaikuad, A, Dixon-Clarke, S, McCorvie, T.J, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2015-09-25 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Ligand-Dependent Dimerization of Phenylalanine Hydroxylase Regulatory Domain.
Sci.Rep., 6, 2016
|
|
4I2P
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with rilpivirine (TMC278) based analogue | | Descriptor: | (2E)-3-[4-({6-[(4-methoxyphenyl)amino]-7H-purin-2-yl}amino)-3,5-dimethylphenyl]prop-2-enenitrile, Gag-Pol polyprotein | | Authors: | Patel, D, Bauman, J.D, Das, K, Arnold, E. | | Deposit date: | 2012-11-22 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2964 Å) | | Cite: | A comparison of the ability of rilpivirine (TMC278) and selected analogues to inhibit clinically relevant HIV-1 reverse transcriptase mutants.
Retrovirology, 9, 2012
|
|
4I2Q
 
 | | Crystal structure of K103N/Y181C mutant of HIV-1 reverse transcriptase in complex with rilpivirine (TMC278) analogue | | Descriptor: | (2E)-3-(4-{[6-(1,3-benzothiazol-5-ylamino)-9H-purin-2-yl]amino}-3,5-dimethylphenyl)prop-2-enenitrile, 1,2-ETHANEDIOL, Gag-Pol polyprotein | | Authors: | Patel, D, Bauman, J.D, Das, K, Arnold, E. | | Deposit date: | 2012-11-22 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7004 Å) | | Cite: | A comparison of the ability of rilpivirine (TMC278) and selected analogues to inhibit clinically relevant HIV-1 reverse transcriptase mutants.
Retrovirology, 9, 2012
|
|
3K94
 
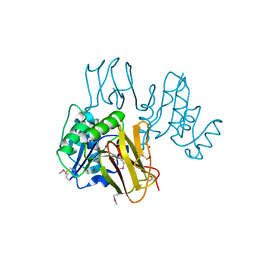 | | Crystal Structure of Thiamin pyrophosphokinase from Geobacillus thermodenitrificans, Northeast Structural Genomics Consortium Target GtR2 | | Descriptor: | Thiamin pyrophosphokinase | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Janjua, J, Xiao, R, Patel, D.J, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Northeast Structural Genomics Consortium Target GtR2
To be Published
|
|
6O6S
 
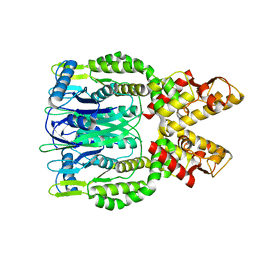 | | Crystal structure of Apo Csm6 | | Descriptor: | Csm6 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4Cleavage with ApA>p Formation Terminating RNase Activity.
Mol.Cell, 75, 2019
|
|
7R76
 
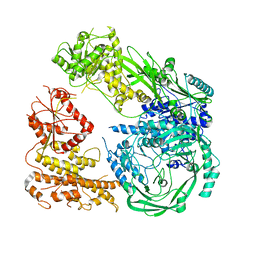 | | Cryo-EM structure of DNMT5 in apo state | | Descriptor: | DNA repair protein Rad8, ZINC ION | | Authors: | Wang, J, Patel, D.J. | | Deposit date: | 2021-06-24 | | Release date: | 2022-02-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into DNMT5-mediated ATP-dependent high-fidelity epigenome maintenance.
Mol.Cell, 82, 2022
|
|
6O74
 
 | |
6O7D
 
 | | Crystal structure of Csm1-Csm4 cassette in complex with one ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), Csm4, ... | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Second Messenger cA4Formation within the Composite Csm1 Palm Pocket of Type III-A CRISPR-Cas Csm Complex and Its Release Path.
Mol.Cell, 75, 2019
|
|
6O7H
 
 | |
6O6X
 
 | |
4QUF
 
 | | crystal structure of chromodomain of Rhino with H3K9me3 | | Descriptor: | H3(1-15)K9me3 peptide, RE36324p | | Authors: | Li, S, Patel, D.J. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing.
Genes Dev., 28, 2014
|
|
4RGF
 
 | | Crystal structure of the in-line aligned env22 twister ribozyme soaked with Mn2+ | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Ren, A, Rajashankar, K.R, Simanshu, D, Patel, D. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2008 Å) | | Cite: | In-line alignment and Mg(2+) coordination at the cleavage site of the env22 twister ribozyme.
Nat Commun, 5, 2014
|
|
4RGE
 
 | | Crystal structure of the in-line aligned env22 twister ribozyme | | Descriptor: | MAGNESIUM ION, env22 twister ribozyme | | Authors: | Ren, A, Rajashankar, K.R, Simanshu, D, Patel, D. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | In-line alignment and Mg(2+) coordination at the cleavage site of the env22 twister ribozyme.
Nat Commun, 5, 2014
|
|
6OV0
 
 | |
2X27
 
 | | Crystal structure of the outer membrane protein OprG from Pseudomonas aeruginosa | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NICKEL (II) ION, OUTER MEMBRANE PROTEIN OPRG | | Authors: | Touw, D.S, Patel, D.R, van den Berg, B. | | Deposit date: | 2010-01-11 | | Release date: | 2010-12-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Oprg from Pseudomonas Aeruginosa, a Potential Channel for Transport of Hydrophobic Molecules Across the Outer Membrane.
Plos One, 5, 2010
|
|
4DOW
 
 | | Structure of mouse ORC1 BAH domain bound to H4K20me2 | | Descriptor: | Histone H4, Origin recognition complex subunit 1 | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2012-02-10 | | Release date: | 2012-03-07 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome.
Nature, 484, 2012
|
|
