1EDQ
 
 | |
1EHN
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT E315Q COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-02-22 | | Release date: | 2001-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
1FFR
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT Y390F COMPLEXED WITH HEXA-N-ACETYLCHITOHEXAOSE (NAG)6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
1FFQ
 
 | | CRYSTAL STRUCTURE OF CHITINASE A COMPLEXED WITH ALLOSAMIDIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITINASE A | | Authors: | Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo purification scheme and crystallization conditions yield high-resolution structures of chitinase A and its complex with the inhibitor allosamidin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1EIB
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT D313A COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-02-25 | | Release date: | 2001-02-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
2WLS
 
 | | Crystal structure of Mus musculus Acetylcholinesterase in complex with AMTS13 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pang, Y.P, Ekstrom, F, Polsinelli, G.A, Gao, Y, Rana, S, Hua, D.H, Andersson, B, Andersson, P.O, Peng, L, Singh, S.K, Mishra, R.K, Zhu, K.Y, Fallon, A.M, Ragsdale, D.W, Brimijoin, S. | | Deposit date: | 2009-06-25 | | Release date: | 2009-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selective and Irreversible Inhibitors of Mosquito Acetylcholinesterases for Controlling Malaria and Other Mosquito-Borne Diseases.
Plos One, 4, 2009
|
|
2GYV
 
 | | Crystal structure of Mus musculus Acetylcholinesterase in complex with Ortho-7 | | Descriptor: | 1,7-HEPTYLENE-BIS-N,N'-SYN-2-PYRIDINIUMALDOXIME, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15-PENTAOXAHEPTADECANE, ... | | Authors: | Pang, Y.P, Boman, M, Artursson, E, Akfur, C, Lundberg, S. | | Deposit date: | 2006-05-10 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of acetylcholinesterase in complex with HI-6, Ortho-7 and obidoxime: Structural basis for differences in the ability to reactivate tabun conjugates.
Biochem.Pharm., 72, 2006
|
|
3J9K
 
 | | Structure of Dark apoptosome in complex with Dronc CARD domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Apaf-1 related killer DARK, Caspase Nc | | Authors: | Pang, Y, Bai, X, Yan, C, Hao, Q, Chen, Z, Wang, J, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-02-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the apoptosome: mechanistic insights into activation of an initiator caspase from Drosophila.
Genes Dev., 29, 2015
|
|
2GYW
 
 | | Crystal Structure of Mus musculus Acetylcholinesterase in Complex with Obidoxime | | Descriptor: | 1,1'-(OXYDIMETHYLENE)BIS(4-FORMYLPYRIDINIUM)DIOXIME, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Pang, Y.P, Boman, M, Artursson, E, Akfur, C, Lundberg, S. | | Deposit date: | 2006-05-10 | | Release date: | 2006-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of acetylcholinesterase in complex with HI-6, Ortho-7 and obidoxime: Structural basis for differences in the ability to reactivate tabun conjugates.
Biochem.Pharm., 72, 2006
|
|
2GYU
 
 | | Crystal structure of Mus musculus Acetylcholinesterase in complex with HI-6 | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ... | | Authors: | Pang, Y.P, Boman, M, Artursson, E, Akfur, C, Lundberg, S. | | Deposit date: | 2006-05-10 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of acetylcholinesterase in complex with HI-6, Ortho-7 and obidoxime: Structural basis for differences in the ability to reactivate tabun conjugates.
Biochem.Pharm., 72, 2006
|
|
2FSI
 
 | |
2FSF
 
 | |
2FSG
 
 | |
2FSH
 
 | |
1HN4
 
 | | PROPHOSPHOLIPASE A2 DIMER COMPLEXED WITH MJ33, SULFATE, AND CALCIUM | | Descriptor: | 1-HEXADECYL-3-TRIFLUOROETHYL-SN-GLYCERO-2-PHOSPHATE METHANE, CALCIUM ION, PROPHOSPHOLIPASE A2, ... | | Authors: | Epstein, T.M, Pan, Y.H, Jain, M.K, Bahnson, B.J. | | Deposit date: | 2000-12-06 | | Release date: | 2001-12-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The basis for k(cat) impairment in prophospholipase A(2) from the anion-assisted dimer structure.
Biochemistry, 40, 2001
|
|
3ZOA
 
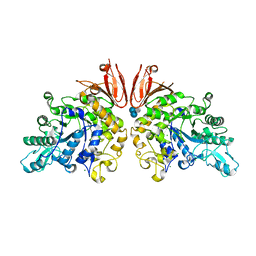 | | The structure of Trehalose Synthase (TreS) of Mycobacterium smegmatis in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Caner, S, Nguyen, N, Aguda, A, Zhang, R, Pan, Y.T, Withers, S.G, Brayer, G.D. | | Deposit date: | 2013-02-21 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of the Mycobacterium Smegmatis Trehalose Synthase Reveals an Unusual Active Site Configuration and Acarbose-Binding Mode.
Glycobiology, 23, 2013
|
|
3ZO9
 
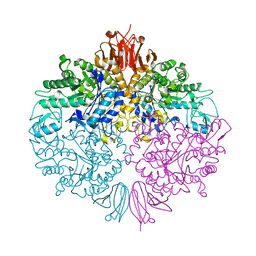 | | The structure of Trehalose Synthase (TreS) of Mycobacterium smegmatis | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Caner, S, Nguyen, N, Aguda, A, Zhang, R, Pan, Y.T, Withers, S.G, Brayer, G.D. | | Deposit date: | 2013-02-21 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Structure of the Mycobacterium Smegmatis Trehalose Synthase Reveals an Unusual Active Site Configuration and Acarbose-Binding Mode.
Glycobiology, 23, 2013
|
|
8DL7
 
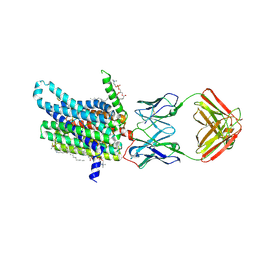 | | Cryo-EM structure of human ferroportin/slc40 bound to minihepcidin PR73 in nanodisc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 11F9 heavy-chain, 11F9 light-chain, ... | | Authors: | Shen, J, Wilbon, A.S, Pan, Y, Zhou, M. | | Deposit date: | 2022-07-07 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of ferroportin inhibition by minihepcidin PR73.
Plos Biol., 21, 2023
|
|
8DL8
 
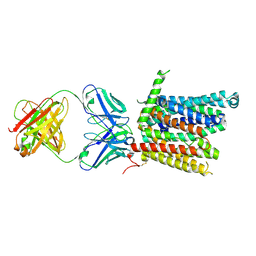 | | Cryo-EM structure of human ferroportin/slc40 bound to Co2+ in nanodisc | | Descriptor: | 11F9 heavy-chain, 11F9 light-chain, COBALT (II) ION, ... | | Authors: | Shen, J, Wilbon, A.S, Pan, Y, Zhou, M. | | Deposit date: | 2022-07-07 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of ferroportin inhibition by minihepcidin PR73.
Plos Biol., 21, 2023
|
|
7XBO
 
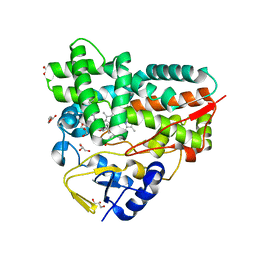 | | Crystal Structure of 10-dml-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | Descriptor: | (3R,4S,5S,7R,9E,11R,12R)-12-ETHYL-4-HYDROXY-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, Cytochrome P450 monooxygenase PikC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | New mechanistic insight of cytochrome P450 PikC gained from site-specific mutagenesis by non-coding amino acids
Nat Commun, 2023
|
|
7XBN
 
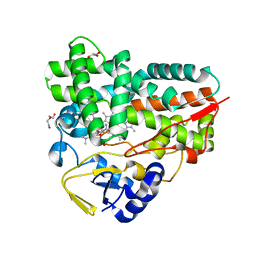 | | Crystal Structure of YC-17-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, Cytochrome P450 monooxygenase PikC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New mechanistic insight of cytochrome P450 PikC gained from site-specific mutagenesis by non-coding amino acids
Nat Commun, 2023
|
|
7XBM
 
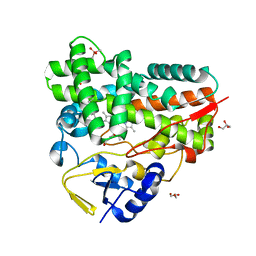 | | Crystal Structure of cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | Descriptor: | CACODYLATE ION, Cytochrome P450 monooxygenase PikC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238
Nat Commun, 2023
|
|
8DL6
 
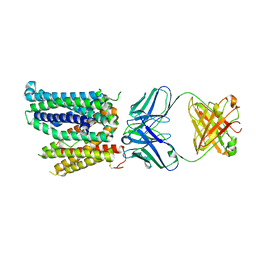 | | Cryo-EM structure of human ferroportin/slc40 bound to Ca2+ in nanodisc | | Descriptor: | 11F9 heavy-chain, 11F9 light-chain, CALCIUM ION, ... | | Authors: | Shen, J, Wilbon, A.S, Pan, Y, Zhou, M. | | Deposit date: | 2022-07-07 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of Ca 2+ transport by ferroportin.
Elife, 12, 2023
|
|
8U27
 
 | | Bcl-2-xL complexed with compound 35 | | Descriptor: | Apoptosis regulator Bcl-2, Bcl-2-like protein 1 chimera, propan-2-yl {4-[(5S)-1-(4-bromobenzoyl)-5-phenyl-4,5-dihydro-1H-pyrazol-3-yl]phenyl}carbamate | | Authors: | Rizo, J, Pan, Y.-Z. | | Deposit date: | 2023-09-05 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights for selective disruption of Beclin 1 binding to Bcl-2.
Commun Biol, 6, 2023
|
|
6VYH
 
 | | Cryo-EM structure of SLC40/ferroportin in complex with Fab | | Descriptor: | 11F9 heavy-chain, 11F9 light-chain, COBALT (II) ION, ... | | Authors: | Shen, J, Ren, Z, Pan, Y, Gao, S, Yan, N, Zhou, M. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-11 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of ion transport and inhibition in ferroportin.
Nat Commun, 11, 2020
|
|
