1T7D
 
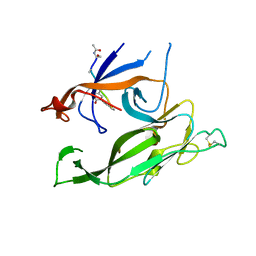 | | Crystal structure of Escherichia coli type I signal peptidase in complex with a lipopeptide inhibitor | | Descriptor: | 10-METHYLUNDECANOIC ACID, ARYLOMYCIN A2, SIGNAL PEPTIDASE I | | Authors: | Paetzel, M, Goodall, J.J, Kania, M, Dalbey, R.E, Page, M.G.P. | | Deposit date: | 2004-05-09 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystallographic and Biophysical Analysis of a Bacterial Signal Peptidase in Complex with a Lipopeptide Based Inhibitor.
J.Biol.Chem., 279, 2004
|
|
3BLC
 
 | |
2PNM
 
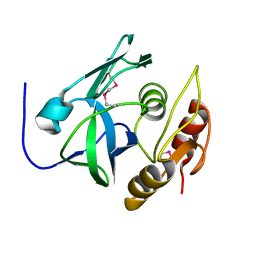 | |
1KN9
 
 | |
1B12
 
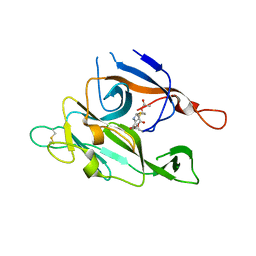 | | CRYSTAL STRUCTURE OF TYPE 1 SIGNAL PEPTIDASE FROM ESCHERICHIA COLI IN COMPLEX WITH A BETA-LACTAM INHIBITOR | | Descriptor: | PHOSPHATE ION, SIGNAL PEPTIDASE I, prop-2-en-1-yl (2S)-2-[(2S,3R)-3-(acetyloxy)-1-oxobutan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylate | | Authors: | Paetzel, M, Dalbey, R, Strynadka, N.C.J. | | Deposit date: | 1999-11-24 | | Release date: | 1999-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor.
Nature, 396, 1998
|
|
2GEF
 
 | |
3IIQ
 
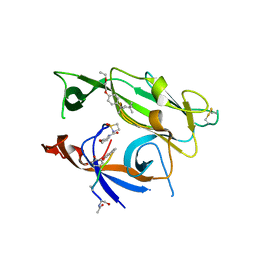 | |
3BEZ
 
 | |
3BF0
 
 | |
4IZJ
 
 | |
4IZK
 
 | |
3TGO
 
 | | Crystal structure of the E. coli BamCD complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Lipoprotein 34, ... | | Authors: | Paetzel, M, Kim, K.H, Aulakh, S. | | Deposit date: | 2011-08-17 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the beta-barrel assembly machinery BamCD protein complex
J.Biol.Chem., 286, 2011
|
|
2PNL
 
 | | Crystal structure of VP4 protease from infectious pancreatic necrosis virus (IPNV) in space group P1 | | Descriptor: | GUANIDINE, Protease VP4 | | Authors: | Paetzel, M, Lee, J, Feldman, A.R, Delmas, B. | | Deposit date: | 2007-04-24 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the VP4 protease from infectious pancreatic necrosis virus reveals the acyl-enzyme complex for an intermolecular self-cleavage reaction.
J.Biol.Chem., 282, 2007
|
|
1FOF
 
 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 | | Descriptor: | BETA LACTAMASE OXA-10, COBALT (II) ION, SULFATE ION | | Authors: | Paetzel, M, Danel, F, de Castro, L, Mosimann, S.C, Page, M.G.P, Strynadka, N.C.J. | | Deposit date: | 2000-08-28 | | Release date: | 2000-10-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the class D beta-lactamase OXA-10.
Nat.Struct.Biol., 7, 2000
|
|
3S04
 
 | |
2Q2I
 
 | | Crystal structure of the protein secretion chaperone CsaA from Agrobacterium tumefaciens. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Secretion chaperone | | Authors: | Feldman, A.R, Shapova, Y.A, Paetzel, M. | | Deposit date: | 2007-05-28 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Phage display and crystallographic analysis reveals potential substrate/binding site interactions in the protein secretion chaperone CsaA from Agrobacterium tumefaciens.
J.Mol.Biol., 379, 2008
|
|
2Q2H
 
 | | Crystal structure of the protein secretion chaperone CsaA from Agrobacterium tumefaciens with a genetically fused phage-display derived peptide substrate at the N-terminus. | | Descriptor: | ACETATE ION, CITRIC ACID, Secretion chaperone, ... | | Authors: | Feldman, A.R, Shapova, Y.A, Paetzel, M. | | Deposit date: | 2007-05-28 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Phage display and crystallographic analysis reveals potential substrate/binding site interactions in the protein secretion chaperone CsaA from Agrobacterium tumefaciens.
J.Mol.Biol., 379, 2008
|
|
3HL4
 
 | | Crystal structure of a mammalian CTP:phosphocholine cytidylyltransferase with CDP-choline | | Descriptor: | Choline-phosphate cytidylyltransferase A, FORMIC ACID, GLYCEROL, ... | | Authors: | Lee, J, Paetzel, M, Cornell, R.B. | | Deposit date: | 2009-05-26 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a mammalian CTP: Phosphocholine cytidylyltransferase catalytic domain reveals novel active site residues within a highly conserved nucleotidyl-transferase fold
J.Biol.Chem., 284, 2009
|
|
8DRX
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp10-nsp11 (C10) cut site sequence (form 2) | | Descriptor: | Fusion protein of 3C-like proteinase nsp5 and nsp10-nsp11 (C10) cut site, SODIUM ION | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRS
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp6-nsp7 (C6) cut site sequence | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRT
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp6-nsp7 (C6) cut site sequence (form 2) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRW
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp9-nsp10 (C9) cut site sequence | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp9-nsp10 (C9) cut site, PENTAETHYLENE GLYCOL, ... | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRU
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp7-nsp8 (C7) cut site sequence | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp7-nsp8 (C7) cut site, PENTAETHYLENE GLYCOL, ... | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRR
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp4-nsp5 (C4) cut site sequence | | Descriptor: | 3C-like proteinase nsp5, SODIUM ION | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRV
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp8-nsp9 (C8) cut site sequence | | Descriptor: | Fusion protein of 3C-like proteinase nsp5 and nsp8-nsp9 (C8) cut site, PENTAETHYLENE GLYCOL | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
