5KE3
 
 | | Crystal structure of SETDB1 Tudor domain in complex with fragment MRT0181a | | Descriptor: | (S)-N-(furan-2-ylmethyl)-1-(1,2,3,4-tetrahydroisoquinoline-3-carbonyl)piperidine-4-carboxamide, BETA-MERCAPTOETHANOL, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Dong, A, Iqbal, A, Mader, P, Dobrovetsky, E, Ferreira de Freitas, R, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Schapira, M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-09 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain in complex with fragment MRT0181a
to be published
|
|
5K4D
 
 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 3 | | Descriptor: | Eukaryotic translation initiation factor 3 subunit D | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
5K77
 
 | | Dbr1 in complex with 7-mer branched RNA | | Descriptor: | FE (II) ION, HYDROXIDE ION, RNA lariat debranching enzyme, ... | | Authors: | Clark, N.E, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Metal dependence and branched RNA cocrystal structures of the RNA lariat debranching enzyme Dbr1.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5JH5
 
 | | Structural Basis for the Hierarchical Assembly of the Core of PRC1.1 | | Descriptor: | BCL-6 corepressor-like protein 1, Lysine-specific demethylase 2B, Polycomb group RING finger protein 1, ... | | Authors: | Wong, S.J, Taylor, A.B, Hart, P.J, Kim, C.A. | | Deposit date: | 2016-04-20 | | Release date: | 2016-09-14 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | KDM2B Recruitment of the Polycomb Group Complex, PRC1.1, Requires Cooperation between PCGF1 and BCORL1.
Structure, 24, 2016
|
|
5JAP
 
 | |
5JAT
 
 | |
5JAR
 
 | |
5K73
 
 | | as-isolated Dbr1 with Fe(II) and Zn(II) | | Descriptor: | FE (II) ION, HYDROXIDE ION, RNA lariat debranching enzyme, ... | | Authors: | Clark, N.E, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The RNA lariat debranching enzyme Dbr1: metal dependence and branched RNA co-crystal structures
Proc.Natl.Acad.Sci.USA, 2016
|
|
5JAL
 
 | |
5JAS
 
 | |
5I9N
 
 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT412 | | Descriptor: | 5-ethyl-4-fluoro-2-(2-nitrophenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.512 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5IGX
 
 | |
5K71
 
 | | apo Dbr1 | | Descriptor: | RNA lariat debranching enzyme, putative, SULFATE ION | | Authors: | Clark, N.E, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The RNA lariat debranching enzyme Dbr1: metal dependence and branched RNA co-crystal structures
Proc.Natl.Acad.Sci.USA, 2016
|
|
5KCJ
 
 | | Structure of the human GluN1/GluN2A LBD in complex with GNE6901 | | Descriptor: | 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-06-06 | | Release date: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Positive Allosteric Modulators of GluN2A-Containing NMDARs with Distinct Modes of Action and Impacts on Circuit Function.
Neuron, 89, 2016
|
|
5KCH
 
 | | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy into weak electron density | | Descriptor: | 4-methoxy-N-[(pyridin-2-yl)methyl]aniline, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Tempel, W, Harding, R.J, Mader, P, Dobrovetsky, E, Walker, J.R, Brown, P.J, Schapira, M, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Santhakumar, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-06 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy
To Be Published
|
|
5KH6
 
 | | SETDB1 in complex with a fragment candidate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Walker, J.R, Harding, R.J, Mader, P, Dobrovetsky, E, Dong, A, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, von Delft, F, Brown, P.J, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Santhakumar, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | SETDB1 in complex with a fragment candidate
To be published
|
|
5KCO
 
 | | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy | | Descriptor: | DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase SETDB1, SULFATE ION, ... | | Authors: | Tempel, W, Harding, R.J, Mader, P, Dobrovetsky, E, Walker, J.R, Brown, P.J, Schapira, M, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Santhakumar, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-06 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy
To Be Published
|
|
5KE2
 
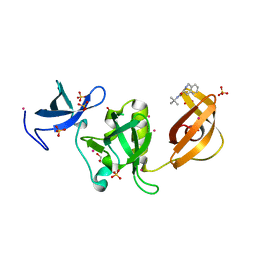 | | Crystal structure of SETDB1 Tudor domain in complex with inhibitor XST06472A | | Descriptor: | (3~{S})-~{N}-~{tert}-butyl-1,2,3,4-tetrahydroisoquinoline-3-carboxamide, 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Dong, A, Iqbal, A, Mader, P, Dobrovetsky, E, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain in complex with inhibitor xst06472a
to be published
|
|
5JAU
 
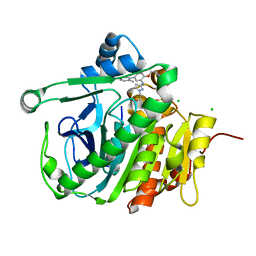 | |
5JAO
 
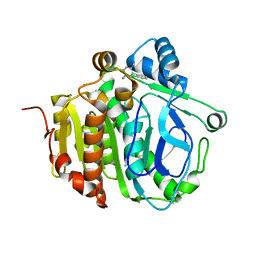 | |
5JJ5
 
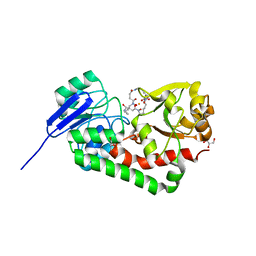 | | Crystal structure of iron uptake ABC transporter substrate-binding protein PiaA from Streptococcus pneumoniae Canada MDR_19A bound to hydroxymate siderophore ferrioxamine E and iron(III) | | Descriptor: | (8E)-6,17,28-trihydroxy-1,6,12,17,23,28-hexaazacyclotritriacont-8-ene-2,5,13,16,24,27-hexone, ABC transporter substrate-binding protein-iron transport, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kurdritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-22 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of iron uptake ABC transporter substrate-binding protein PiaA from Streptococcus pneumoniae Canada MDR_19A bound to hydroxymate siderophore ferrioxamine E and iron(III)
To Be Published
|
|
5IFL
 
 | | Crystal structure of B. pseudomallei FabI in complex with NAD and triclosan | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5I4A
 
 | | X-ray crystal structure of Marinitoga piezophila Argonaute in complex with 5' OH guide RNA | | Descriptor: | Argonaute protein, RNA (5'-R(*UP*AP*UP*AP*CP*AP*AP*CP*CP*UP*AP*CP*UP*U)-3') | | Authors: | Doxzen, K.W, Kaya, E, Knoll, K.R, Wilson, R.C, Strutt, S.C, Kranzusch, P.J, Doudna, J.A. | | Deposit date: | 2016-02-11 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | A bacterial Argonaute with noncanonical guide RNA specificity.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5KOC
 
 | | Pavine N-methyltransferase in complex with S-adenosylmethionine pH 7 | | Descriptor: | Pavine N-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Torres, M.A, Hoffarth, E, Savtchouk, J, Chen, X, Morris, J, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2016-06-30 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism.
J.Biol.Chem., 291, 2016
|
|
5KOK
 
 | | Pavine N-methyltransferase in complex with Tetrahydropapaverine and S-adenosylhomocysteine pH 7.25 | | Descriptor: | (1~{R})-1-[(3,4-dimethoxyphenyl)methyl]-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline, (1~{S})-1-[(3,4-dimethoxyphenyl)methyl]-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline, Pavine N-methyltransferase, ... | | Authors: | Torres, M.A, Hoffarth, E, Eugenio, L, Savtchouk, J, Chen, X, Morris, J, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2016-06-30 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism.
J.Biol.Chem., 291, 2016
|
|
