1XD3
 
 | | Crystal structure of UCHL3-UbVME complex | | 分子名称: | MAGNESIUM ION, METHYL 4-AMINOBUTANOATE, UBC protein, ... | | 著者 | Misaghi, S, Galardy, P.J, Meester, W.J.N, Ovaa, H, Ploegh, H.L, Gaudet, R. | | 登録日 | 2004-09-03 | | 公開日 | 2004-11-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure of the Ubiquitin Hydrolase UCH-L3 Complexed with a Suicide Substrate
J.Biol.Chem., 280, 2005
|
|
1XBO
 
 | | PTP1B complexed with Isoxazole Carboxylic Acid | | 分子名称: | 5-(3-{3-[3-HYDROXY-2-(METHOXYCARBONYL)PHENOXY]PROPENYL}PHENYL)-4-(HYDROXYMETHYL)ISOXAZOLE-3-CARBOXYLIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | 著者 | Zhao, H, Liu, G, Xin, Z, Serby, M, Pei, Z, Szczepankiewicz, B.G, Hajduk, P.J, Abad-Zapatero, C, Hutchins, C.W, Lubben, T.H, Ballaron, S.J, Hassach, D.L, Kaszubska, W, Rondinone, C.M, Trevillyan, J.M, Jirousek, M.R. | | 登録日 | 2004-08-31 | | 公開日 | 2004-10-19 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Isoxazole carboxylic acids as protein tyrosine phosphatase 1B (PTP1B) inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
6MN0
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, H168A mutant in complex with acetyl-CoA | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETYL COENZYME *A, Aminoglycoside N(3)-acetyltransferase, ... | | 著者 | Stogios, P.J, Skarina, T, Zu, X, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-10-01 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
6MN2
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with sisomicin-CoA | | 分子名称: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, ... | | 著者 | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-10-01 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.744 Å) | | 主引用文献 | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with sisomicin-CoA
To Be Published
|
|
6MR3
 
 | | Crystal structure of the competence-damaged protein (CinA) superfamily protein from Streptococcus mutans | | 分子名称: | CHLORIDE ION, Putative competence-damage inducible protein | | 著者 | Stogios, P.J, Cuff, M, Xu, X, Cui, H, Di Leo, R, Yim, V, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2018-10-11 | | 公開日 | 2018-10-24 | | 最終更新日 | 2020-05-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structure of the competence-damaged protein (CinA) superfamily protein from Streptococcus mutans
To Be Published
|
|
6MO6
 
 | | Crystal structure of the selenomethionine-substituted human sulfide:quinone oxidoreductase | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Sulfide:quinone oxidoreductase, ... | | 著者 | Jackson, M.R, Jorns, M.S, Loll, P.J. | | 登録日 | 2018-10-04 | | 公開日 | 2019-04-10 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | X-Ray Structure of Human Sulfide:Quinone Oxidoreductase: Insights into the Mechanism of Mitochondrial Hydrogen Sulfide Oxidation.
Structure, 27, 2019
|
|
1XK9
 
 | | Pseudomanas exotoxin A in complex with the PJ34 inhibitor | | 分子名称: | Exotoxin A, N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE | | 著者 | Yates, S.P, Taylor, P.J, Joergensen, R, Ferrraris, D, Zhang, J, Andersen, G.R, Merrill, A.R. | | 登録日 | 2004-09-28 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-function analysis of water-soluble inhibitors of the catalytic domain of exotoxin A from Pseudomonas aeruginosa
BIOCHEM.J., 385, 2005
|
|
7QFU
 
 | | Crystal Structure of AtlA catalytic domain from Enterococcus feacalis | | 分子名称: | GLYCEROL, Peptidoglycan hydrolase | | 著者 | Zamboni, V, Barelier, S, Dixon, R, Galley, N, Ghanem, A, Cahuzac, H, Salamaga, B, Davis, P.J, Mesnage, S, Vincent, F. | | 登録日 | 2021-12-06 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Molecular basis for substrate recognition and septum cleavage by AtlA, the major N-acetylglucosaminidase of Enterococcus faecalis.
J.Biol.Chem., 298, 2022
|
|
1XF9
 
 | | Structure of NBD1 from murine CFTR- F508S mutant | | 分子名称: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, ... | | 著者 | Thibodeau, P.H, Brautigam, C.A, Machius, M, Thomas, P.J. | | 登録日 | 2004-09-14 | | 公開日 | 2004-12-28 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Side chain and backbone contributions of Phe508 to CFTR folding.
Nat.Struct.Mol.Biol., 12, 2005
|
|
6MYD
 
 | | Structure of zebrafish TRAF6 in complex with STING CTT | | 分子名称: | STING CTT, Transmembrane protein 173, SULFATE ION, ... | | 著者 | de Oliveira Mann, C.C, Orzalli, M.H, King, D.S, Kagan, J.C, Lee, A.S.Y, Kranzusch, P.J. | | 登録日 | 2018-11-01 | | 公開日 | 2019-05-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.399 Å) | | 主引用文献 | Modular Architecture of the STING C-Terminal Tail Allows Interferon and NF-kappa B Signaling Adaptation.
Cell Rep, 27, 2019
|
|
1CDR
 
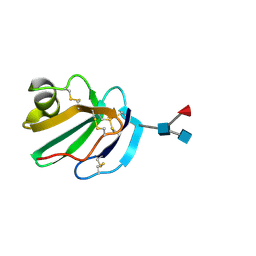 | | STRUCTURE OF A SOLUBLE, GLYCOSYLATED FORM OF THE HUMAN COMPLEMENT REGULATORY PROTEIN CD59 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CD59 | | 著者 | Fletcher, C.M, Harrison, R.A, Lachmann, P.J, Neuhaus, D. | | 登録日 | 1994-06-01 | | 公開日 | 1994-09-30 | | 最終更新日 | 2020-07-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of a soluble, glycosylated form of the human complement regulatory protein CD59.
Structure, 2, 1994
|
|
6N4G
 
 | |
1S9G
 
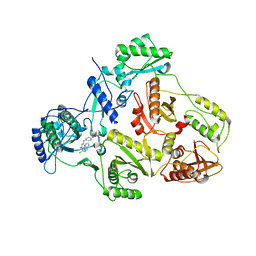 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R120394. | | 分子名称: | 4-[4-AMINO-6-(5-CHLORO-1H-INDOL-4-YLMETHYL)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | 著者 | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | 登録日 | 2004-02-04 | | 公開日 | 2004-05-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1S1J
 
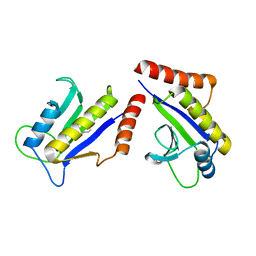 | | Crystal Structure of ZipA in complex with indoloquinolizin inhibitor 1 | | 分子名称: | (12bS)-1,2,3,4,12,12b-hexahydroindolo[2,3-a]quinolizin-7(6H)-one, Cell division protein zipA | | 著者 | Jenning, L.D, Foreman, K.W, Rush III, T.S, Tsao, D.H, Mosyak, L, Li, Y, Sukhdeo, M.N, Ding, W, Dushin, E.G, Kenney, C.H, Moghazeh, S.L, Peterson, P.J, Ruzin, A.V, Tuckman, M, Sutherland, A.G. | | 登録日 | 2004-01-06 | | 公開日 | 2004-05-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Design and synthesis of indolo[2,3-a]quinolizin-7-one inhibitors of the ZipA-FtsZ interaction
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1CDQ
 
 | | STRUCTURE OF A SOLUBLE, GLYCOSYLATED FORM OF THE HUMAN COMPLEMENT REGULATORY PROTEIN CD59 | | 分子名称: | CD59 | | 著者 | Fletcher, C.M, Harrison, R.A, Lachmann, P.J, Neuhaus, D. | | 登録日 | 1994-06-01 | | 公開日 | 1994-09-30 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of a soluble, glycosylated form of the human complement regulatory protein CD59.
Structure, 2, 1994
|
|
7QCP
 
 | |
1YRS
 
 | | Crystal structure of KSP in complex with inhibitor 1 | | 分子名称: | 3-[(5S)-1-ACETYL-3-(2-CHLOROPHENYL)-4,5-DIHYDRO-1H-PYRAZOL-5-YL]PHENOL, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | 著者 | Cox, C.D, Breslin, M.J, Mariano, B.J, Coleman, P.J, Buser, C.A, Walsh, E.S, Hamilton, K, Kohl, N.E, Torrent, M, Yan, Y, Kuo, L.C, Hartman, G.D. | | 登録日 | 2005-02-04 | | 公開日 | 2005-04-12 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Kinesin spindle protein (KSP) inhibitors. Part 1: The discovery of 3,5-diaryl-4,5-dihydropyrazoles as potent and selective inhibitors of the mitotic kinesin KSP
BIOORG.MED.CHEM.LETT., 15, 2005
|
|
7QIB
 
 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 8.5 in complex with UDP | | 分子名称: | CHLORIDE ION, Glucosyl-3-phosphoglycerate synthase, MAGNESIUM ION, ... | | 著者 | Silva, A, Nunes-Costa, D, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | 登録日 | 2021-12-14 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 8.5 in complex with UDP
To Be Published
|
|
7QI9
 
 | |
7QOQ
 
 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 8.5 in complex with UMP and Magnesium | | 分子名称: | CHLORIDE ION, Glucosyl-3-phosphoglycerate synthase, MAGNESIUM ION, ... | | 著者 | Silva, A, Barbosa Pereira, P.J, Macedo-Ribeiro, S, Costa, D. | | 登録日 | 2021-12-27 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 8.5 in complex with UMP and Magnesium
To Be Published
|
|
5SVA
 
 | | Mediator-RNA Polymerase II Pre-Initiation Complex | | 分子名称: | 108bp HIS4 Promoter Non-template Strand (-92/+16), 108bp HIS4 Promoter Template Strand (+16/-92), DNA repair helicase RAD25, ... | | 著者 | Robinson, P.J, Bushnell, D.A, Kornberg, R.D. | | 登録日 | 2016-08-05 | | 公開日 | 2016-09-28 | | 最終更新日 | 2019-11-27 | | 実験手法 | ELECTRON MICROSCOPY (15.3 Å) | | 主引用文献 | Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex.
Cell, 166, 2016
|
|
1YSN
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL complexed with an acyl-sulfonamide-based ligand | | 分子名称: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-X | | 著者 | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | 登録日 | 2005-02-08 | | 公開日 | 2005-06-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1Z6O
 
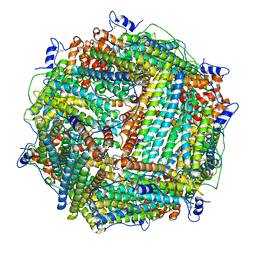 | | Crystal Structure of Trichoplusia ni secreted ferritin | | 分子名称: | CALCIUM ION, FE (III) ION, Ferritin heavy chain, ... | | 著者 | Hamburger, A.E, West Jr, A.P, Hamburger, Z.A, Hamburger, P, Bjorkman, P.J. | | 登録日 | 2005-03-22 | | 公開日 | 2005-05-24 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Crystal structure of a secreted insect ferritin reveals a symmetrical arrangement of heavy and light chains.
J.Mol.Biol., 349, 2005
|
|
7QA1
 
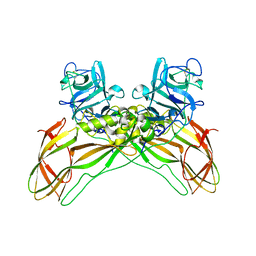 | | The structure of natural crystals of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated using serial femtosecond crystallography at an X-ray free electron laser | | 分子名称: | Toxin-10 pesticidal protein (Tpp) 49Aa1 | | 著者 | Williamson, L.J, Rizkallah, P.J, Berry, C, Oberthur, D, Galchenkova, M, Yefanov, O, Bean, R, Best, H.L. | | 登録日 | 2021-11-15 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated from natural crystals using MHz-SFX.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6NPS
 
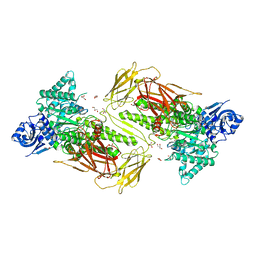 | | Crystal structure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus | | 分子名称: | AxyAgu115A, CHLORIDE ION, GLYCEROL | | 著者 | Stogios, P.J, Skarina, T, Di Leo, R, Yan, R, Master, E, Savchenko, A. | | 登録日 | 2019-01-18 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural characterization of the family GH115 alpha-glucuronidase from Amphibacillus xylanus yields insight into its coordinated action with alpha-arabinofuranosidases.
N Biotechnol, 2021
|
|
