4KGD
 
 | |
5C0D
 
 | | HLA-A02 carrying AQWGPDPAAA | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2015-06-12 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J.Clin.Invest., 126, 2016
|
|
5VM5
 
 | | Engineered tryptophan synthase b-subunit from Pyrococcus furiosus, PfTrpB2B9, with Ser bound | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, SODIUM ION, Tryptophan synthase beta chain 1, ... | | Authors: | Buller, A.R, van Roye, P. | | Deposit date: | 2017-04-26 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Directed Evolution Mimics Allosteric Activation by Stepwise Tuning of the Conformational Ensemble.
J. Am. Chem. Soc., 140, 2018
|
|
5WCD
 
 | | Crystal structure of the broadly neutralizing Influenza A antibody VRC 315 04-1D02 Fab. | | Descriptor: | PHOSPHATE ION, SULFATE ION, VRC315 04-1D02 Fab Heavy chain, ... | | Authors: | Joyce, M.G, Andrews, S.F, Mascola, J.R, McDermott, A.B, Kwong, P.D. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-23 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (1.814 Å) | | Cite: | Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans.
Sci Immunol, 2, 2017
|
|
1R3X
 
 | | INTRAMOLECULAR DNA TRIPLEX WITH RNA THIRD STRAND, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*TP*CP*T)-3'), RNA (5'-R(*UP*CP*UP*CP*UP*CP*UP*U)-3') | | Authors: | Gotfredsen, C.H, Schultze, P, Feigon, J. | | Deposit date: | 1998-02-06 | | Release date: | 1998-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of an Intramolecular Pyrimidine-Purine-Pyrimidine Triplex Containing an RNA Third Strand
J.Am.Chem.Soc., 120, 1998
|
|
5C18
 
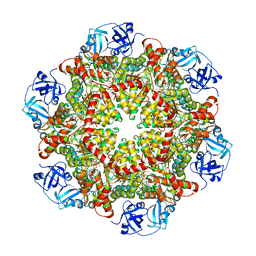 | | p97-delta709-728 in complex with ATP-gamma-S | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2015-06-13 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of ATP Hydrolysis and Intersubunit Signaling in the AAA+ ATPase p97.
Structure, 24, 2016
|
|
7TYL
 
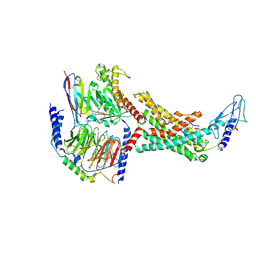 | | Calcitonin Receptor in complex with Gs and rat amylin peptide, bypass motif | | Descriptor: | Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-13 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
3OQ0
 
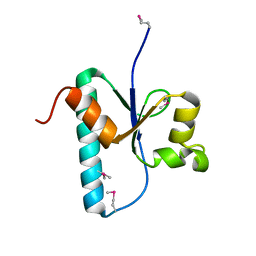 | |
7TYH
 
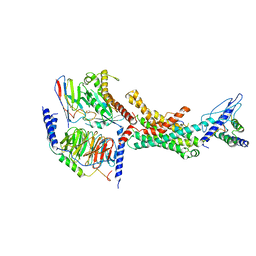 | | Human Amylin2 Receptor in complex with Gs and human calcitonin peptide | | Descriptor: | Calcitonin, Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-13 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
7TYO
 
 | | Calcitonin receptor in complex with Gs and human calcitonin peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Calcitonin, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
3BXS
 
 | | Crystal Structures Of Highly Constrained Substrate And Hydrolysis Products Bound To HIV-1 Protease. Implications For Catalytic Mechanism | | Descriptor: | (9S,12S)-9-(1-methylethyl)-7,10-dioxo-2-oxa-8,11-diazabicyclo[12.2.2]octadeca-1(16),14,17-triene-12-carboxylic acid, Protease, SULFATE ION | | Authors: | Tyndall, J.D, Pattenden, L.K, Reid, R.C, Hu, S.H, Alewood, D, Alewood, P.F, Walsh, T, Fairlie, D.P, Martin, J.L. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Highly Constrained Substrate and Hydrolysis Products Bound to HIV-1 Protease. Implications for the Catalytic Mechanism
Biochemistry, 47, 2008
|
|
4R4D
 
 | | Racemic crystal structure of a magnesium-bound B-DNA duplex | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3', MAGNESIUM ION, SODIUM ION | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Huc, I. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Racemic DNA crystallography.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6FRA
 
 | |
3OY8
 
 | |
6FMK
 
 | | pVHL:EloB:EloC in complex with N-((S)-1-((2S,4R)-4-hydroxy-2-((4-(4-methylthiazol-5-yl)benzyl)carbamothioyl) pyrrolidin-1-yl)-1-thioxopropan-2-yl)acetamide (ligand 4) | | Descriptor: | Elongin-B, Elongin-C, von Hippel-Lindau disease tumor suppressor, ... | | Authors: | Soares, P, Lucas, X, Ciulli, A. | | Deposit date: | 2018-01-31 | | Release date: | 2018-04-11 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Thioamide substitution to probe the hydroxyproline recognition of VHL ligands.
Bioorg. Med. Chem., 26, 2018
|
|
3OYW
 
 | |
6C8Q
 
 | | Crystal structure of NAD synthetase (NadE) from Enterococcus faecalis in complex with NAD+ | | Descriptor: | NH(3)-dependent NAD(+) synthetase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Stogios, P.J, Skarina, T, McChesney, C, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-25 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | To be published
To Be Published
|
|
7TYW
 
 | | Human Amylin1 Receptor in complex with Gs and salmon calcitonin peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Calcitonin receptor, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
6TI4
 
 | | SHMT from Streptococcus thermophilus Tyr55Ser variant in complex with PLP/D-Serine/Lys230 gem diamine complex | | Descriptor: | (2~{R})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-3-oxidanyl-propanoic acid, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Petrillo, G, Hernandez, K, Bujons, J, Clapes, P, Uson, I. | | Deposit date: | 2019-11-21 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of Y55S Serine Hydroxymethyltransferase variant from Streptococcus thermophilus in complex with gem-diamine intermediate of D-serine
To Be Published
|
|
6WVS
 
 | | Hyperstable de novo TIM barrel variant DeNovoTIM15 | | Descriptor: | DeNovoTIM15 hyperstable de novo TIM barrel | | Authors: | Bick, M.J, Haydon, I.C, Caldwell, S.J, Zeymer, C, Huang, P, Fernandez-Velasco, D.A, Baker, D. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Tight and specific lanthanide binding in a de novo TIM barrel with a large internal cavity designed by symmetric domain fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7U0D
 
 | |
3P2X
 
 | | Insulin fibrillation is the Janus face of induced fit. A chiaral clamp stabilizes the native state at the expense of activity | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Hua, Q.X, Wan, Z.L, Huang, K, Hu, S.Q, Phillip, N.F, Jia, W.H, Whittingham, J, Dodson, G.G, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2010-10-04 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity
To be Published
|
|
7LOX
 
 | | The structure of Agmatinase from E. Coli at 3.2 A displaying guanidine in the active site | | Descriptor: | Agmatinase, GUANIDINE, MANGANESE (II) ION | | Authors: | Maturana, P, Figueroa, M, Gonzalez-Ordenes, F, Villalobos, P, Martinez-Oyanedel, J, Uribe, E.A, Castro-Fernandez, V. | | Deposit date: | 2021-02-11 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of Escherichia coli Agmatinase: Catalytic Mechanism and Residues Relevant for Substrate Specificity.
Int J Mol Sci, 22, 2021
|
|
6GLR
 
 | | Crystal structure of hMTH1 N33G in complex with TH scaffold 1 in the presence of acetate | | Descriptor: | 4-phenylpyrimidin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Eberle, S.A, Wiedmer, L, Sledz, P, Caflisch, A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | hMTH1 N33G in complex with TH scaffold 1
To Be Published
|
|
4A3J
 
 | | RNA Polymerase II initial transcribing complex with a 2nt DNA-RNA hybrid and soaked with GMPCPP | | Descriptor: | 5'-D(*AP*GP*CP*TP*AP*GP*CP*TP*TP*TP*CP*BRUP*AP*CP*CP *TP*GP*AP*AP*CP*AP*AP*CP*TP*AP*AP*CP)-3', 5'-D(*GP*TP*AP*GP*AP*AP*AP*GP*CP*TP*AP*GP*CP*TP)-3', 5'-R(*CP*AP)-3', ... | | Authors: | Cheung, A.C.M, Sainsbury, S, Cramer, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis of Initial RNA Polymerase II Transcription.
Embo J., 30, 2011
|
|
