4TPU
 
 | | CRYSTAL STRUCTURE OF FERREDOXIN-DEPENDENT DISULFIDE REDUCTASE FROM METHANOSARCINA ACETIVORANS | | Descriptor: | BROMIDE ION, FE (III) ION, IRON/SULFUR CLUSTER, ... | | Authors: | Kumar, A.K, Yennawar, H.P, Yennawar, N.H, Ferry, J.G. | | Deposit date: | 2014-06-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.355 Å) | | Cite: | Structural and Biochemical Characterization of a Ferredoxin:Thioredoxin Reductase-like Enzyme from Methanosarcina acetivorans.
Biochemistry, 54, 2015
|
|
2VAH
 
 | | Solution structure of a B-DNA hairpin at low pressure. | | Descriptor: | 5'-D(*AP*GP*GP*AP*TP*CP*CP*TP*UP*TP *TP*GP*GP*AP*TP*CP*CP*T)-3' | | Authors: | Williamson, M.P, Wilton, D.J, Ghosh, M, Chary, K.V.A, Akasaka, K. | | Deposit date: | 2007-08-31 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Change in a B-DNA Helix with Hydrostatic Pressure
Nucleic Acids Res., 36, 2008
|
|
2UZ5
 
 | | Solution structure of the fkbp-domain of Legionella pneumophila Mip | | Descriptor: | MACROPHAGE INFECTIVITY POTENTIATOR | | Authors: | Ceymann, A, Horstmann, M, Ehses, P, Schweimer, K, Steinert, M, Kamphausen, T, Fischer, G, Hacker, J, Rosch, P, Faber, C. | | Deposit date: | 2007-04-25 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Domain Motions of the Mip Protein from Legionella Pneumophila
Biochemistry, 45, 2006
|
|
8HVI
 
 | | Activation mechanism of GPR132 by compound NOX-6-7 | | Descriptor: | 3-methyl-5-[(4-oxidanylidene-4-phenyl-butanoyl)amino]-1-benzofuran-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-26 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
5LLM
 
 | | Structure of the thermostabilized EAAT1 cryst mutant in complex with L-ASP and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, ASPARTIC ACID, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J, Assal, R, Legrand, P, Reyes, N. | | Deposit date: | 2016-07-27 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and allosteric inhibition of excitatory amino acid transporter 1.
Nature, 544, 2017
|
|
8EX0
 
 | |
5AFU
 
 | | Cryo-EM structure of dynein tail-dynactin-BICD2N complex | | Descriptor: | ACTIN, CYTOPLASMIC 1, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Urnavicius, L, Zhang, K, Diamant, A.G, Motz, C, Schlager, M.A, Yu, M, Patel, N.A, Robinson, C.V, Carter, A.P. | | Deposit date: | 2015-01-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | The Structure of the Dynactin Complex and its Interaction with Dynein.
Science, 347, 2015
|
|
3IYD
 
 | | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, DNA (98-MER), ... | | Authors: | Hudson, B.P, Quispe, J, Lara, S, Kim, Y, Berman, H, Arnold, E, Ebright, R.H, Lawson, C.L. | | Deposit date: | 2009-08-01 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (19.799999 Å) | | Cite: | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5EGQ
 
 | |
5EHN
 
 | | mAChE-syn TZ2PA5 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-phenyl-5-[5-[3-[2-(1,2,3,4-tetrahydroacridin-9-ylamino)ethyl]-1,2,3-triazol-4-yl]pentyl]phenanthridin-5-ium-3,8-diamine, Acetylcholinesterase, ... | | Authors: | Bourne, Y, Marchot, P. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Steric and Dynamic Parameters Influencing In Situ Cycloadditions to Form Triazole Inhibitors with Crystalline Acetylcholinesterase.
J.Am.Chem.Soc., 138, 2016
|
|
6T6N
 
 | | Crystal structure of Klebsiella pneumoniae FabG2(NADH-dependent) in complex with NADH at 2.5 A resolution | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-[acyl-carrier protein] reductase, D-MALATE, ... | | Authors: | Vella, P, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
7XGS
 
 | | Short vegetative phase protein | | Descriptor: | 1,2-ETHANEDIOL, MADS-box protein SVP | | Authors: | Liao, Y.-T, Wang, H.-C, Ko, T.-P. | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The PHYL1 effector of peanut witches broom (PnWB) phytoplasma alters the miR156/157-mediated squamosa promoter binding protein like regulation and gibberellic acid generation to promote anthocyanin biosynthesis
To Be Published
|
|
8HQE
 
 | | Cryo-EM structure of the apo-GPR132-Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
6C8Z
 
 | | Last common ancestor of ADP-dependent phosphofructokinases from Methanosarcinales | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADP-dependent phosphofructokinase, MAGNESIUM ION, ... | | Authors: | Castro-Fernandez, V, Gonzalez-Ordenes, F, Munoz, S, Fuentes, N, Leonardo, D, Fuentealba, M, Herrera-Morande, A, Maturana, P, Villalobos, P, Garratt, R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | ADP-Dependent Kinases From the Archaeal OrderMethanosarcinalesAdapt to Salt by a Non-canonical Evolutionarily Conserved Strategy.
Front Microbiol, 9, 2018
|
|
5LSO
 
 | |
3IXT
 
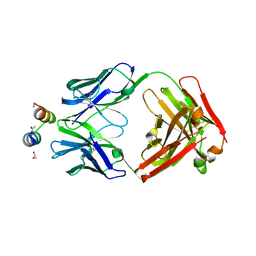 | | Crystal Structure of Motavizumab Fab Bound to Peptide Epitope | | Descriptor: | 1,2-ETHANEDIOL, Fusion glycoprotein F1, Motavizumab Fab heavy chain, ... | | Authors: | McLellan, J.S, Chen, M, Kim, A, Yang, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2009-09-04 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of respiratory syncytial virus neutralization by motavizumab.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5F88
 
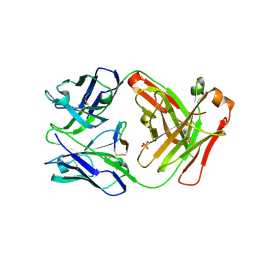 | | Cetuximab Fab in complex with L5Y meditope variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cetuximab Fab heavy chain, Cetuximab Fab light chain, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2015-12-09 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
8HQM
 
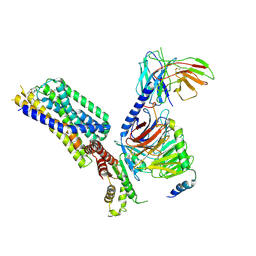 | | Activation mechanism of GPR132 by NPGLY | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
4OSP
 
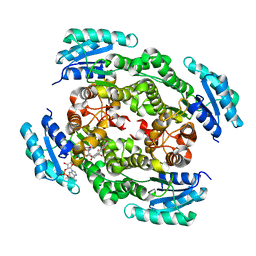 | | The crystal structure of urdamycin C-6 ketoreductase domain UrdMred with bound NADP and rabelomycin | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxygenase-reductase, rabelomycin | | Authors: | Niiranen, L, Paananen, P, Patrikainen, P, Metsa-Ketela, M. | | Deposit date: | 2014-02-13 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based engineering of angucyclinone 6-ketoreductases.
Chem.Biol., 21, 2014
|
|
8EQS
 
 | | Structure of SARS-CoV-1 Orf3a in late endosome/lysosome-like environment, MSP1D1 nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Apolipoprotein A-I, ORF3a protein | | Authors: | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
8HBG
 
 | | FMDV (A/TUR/14/98) in complex with M678F | | Descriptor: | M678F nab, VP1 of capsid protein, VP2 of capsid protein, ... | | Authors: | Li, H.Z, Dong, H, Liu, P. | | Deposit date: | 2022-10-28 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and in vivo studies of neutralizing antibody topographical classifications reveal mechanisms underlying differences in immunogenicity and antigenicity between 146S and 12S of foot-and-mouth disease virus
To Be Published
|
|
6HYS
 
 | | Crystal structure of DHX8 helicase domain bound to ADP at 2.6 angstrom | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Felisberto-Rodrigues, C, Thomas, J.C, McAndrew, P.C, Le Bihan, Y.V, Burke, R, Workman, P, van Montfort, R.L.M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterisation of human RNA helicase DHX8 provides insights into the mechanism of RNA-stimulated ADP release.
Biochem.J., 476, 2019
|
|
2UUM
 
 | | Crystal structure of C-phycocyanin from Phormidium, Lyngbya spp. (Marine) and Spirulina sp. (Fresh water) shows two different ways of energy transfer between two hexamers. | | Descriptor: | BILIVERDINE IX ALPHA, C-PHYCOCYANIN ALPHA CHAIN, C-PHYCOCYANIN BETA CHAIN, ... | | Authors: | Satyanarayana, L, Patel, A, Mishra, S, K Ghosh, P, Suresh, C.G. | | Deposit date: | 2007-03-04 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of C-Phycocyanin from Phormidium, Lyngbya Spp. (Marine) and Spirulina Sp. (Fresh Water) Shows Two Different Ways of Energy Transfer between Two Hexamers.
To be Published
|
|
5LQ0
 
 | | Crystal structure of Tyr24 phosphorylated Annexin A2 at 2.9 A resolution | | Descriptor: | Annexin A2, CALCIUM ION | | Authors: | Ecsedi, P, Gogl, G, Kiss, B, Nyitray, L. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Regulation of the Equilibrium between Closed and Open Conformations of Annexin A2 by N-Terminal Phosphorylation and S100A4-Binding.
Structure, 25, 2017
|
|
6TTD
 
 | | Crystal structure of McoA multicopper oxidase 2F4 variant from the hyperthermophile Aquifex aeolicus | | Descriptor: | COPPER (II) ION, GLYCEROL, Multicopper oxidase, ... | | Authors: | Borges, P.T, Brissos, V, Cordeiro, T.N, Martins, L.O, Frazao, C. | | Deposit date: | 2019-12-26 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Distal Mutations Shape Substrate-Binding Sites during Evolution of a Metallo-Oxidase into a Laccase
Acs Catalysis, 2022
|
|
