1C8Y
 
 | | Endo-Beta-N-Acetylglucosaminidase H, D130A Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H, ZINC ION | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1C9Q
 
 | |
7P18
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Sterolibacterium denitrificans in complex with 1,4-androstadiene-3,17-dione | | Descriptor: | 3-oxosteroid 1-dehydrogenase, ANDROSTA-1,4-DIENE-3,17-DIONE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wojcik, P, Mrugala, B, Kurpiewska, K, Szaleniec, M. | | Deposit date: | 2021-07-01 | | Release date: | 2021-07-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure, Mutagenesis, and QM:MM Modeling of 3-Ketosteroid Delta 1 -Dehydrogenase from Sterolibacterium denitrificans ─The Role of a New Putative Membrane-Associated Domain and Proton-Relay System in Catalysis.
Biochemistry, 62, 2023
|
|
6ZED
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound F1 | | Descriptor: | 1,2-ETHANEDIOL, 4,5-dimethyl-1~{H}-pyrazolo[3,4-c]pyridazin-3-amine, Bromodomain-containing protein 4 | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound F1
To Be Published
|
|
1C7M
 
 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE REDUCED STATE | | Descriptor: | HEME C, PROTEIN (CYTOCHROME C552) | | Authors: | Pristovsek, P, Luecke, C, Reincke, B, Ludwig, B, Rueterjans, H. | | Deposit date: | 2000-03-03 | | Release date: | 2000-07-19 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the functional domain of Paracoccus denitrificans cytochrome c552 in the reduced state.
Eur.J.Biochem., 267, 2000
|
|
7P7A
 
 | | SARS-CoV-2 spike protein in complex with sybody#68 in a 2up/1flexible conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.76 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P77
 
 | | SARS-CoV-2 spike protein in complex with sybody#15 and sybody#68 in a 3up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P79
 
 | | SARS-CoV-2 spike protein in complex with sybodyb#15 in a 1up/1up-out/1down conformation. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P7B
 
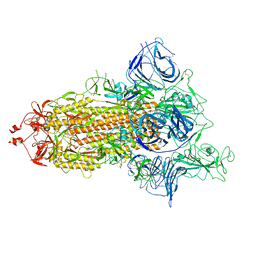 | | SARS-CoV-2 spike protein in complex with sybody no68 in a 1up/2down conformation | | Descriptor: | Spike glycoprotein | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P78
 
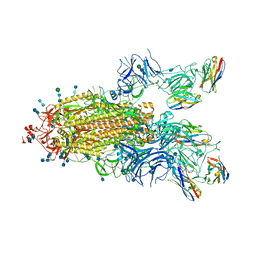 | | SARS-CoV-2 spike protein in complex with sybody#15 and sybody#68 in a 1up/1up-out/1down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7OUG
 
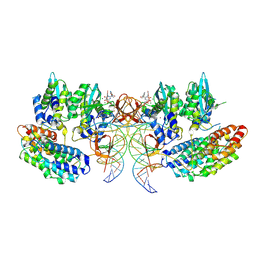 | | STLV-1 intasome:B56 in complex with the strand-transfer inhibitor raltegravir | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), Integrase, ... | | Authors: | Barski, M.S, Ballandras-Colas, A, Cronin, N.B, Pye, V.E, Cherepanov, P, Maertens, G.N. | | Deposit date: | 2021-06-11 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures.
Nat Commun, 12, 2021
|
|
7OUH
 
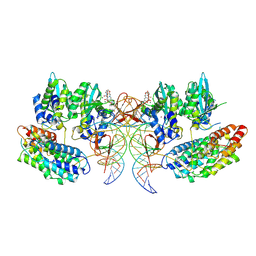 | | Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor bictegravir | | Descriptor: | Bictegravir, DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), ... | | Authors: | Barski, M.S, Ballandras-Colas, A, Cronin, N.B, Pye, V.E, Cherepanov, P, Maertens, G.N. | | Deposit date: | 2021-06-11 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures.
Nat Commun, 12, 2021
|
|
7OUF
 
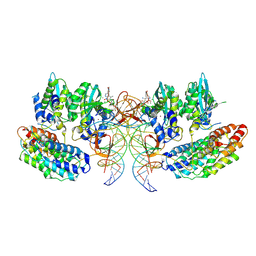 | | Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor XZ450 | | Descriptor: | 4-azanyl-~{N}-[[2,4-bis(fluoranyl)phenyl]methyl]-6-[3-(dimethylamino)-3-oxidanylidene-propyl]-1-oxidanyl-2-oxidanylidene-1,8-naphthyridine-3-carboxamide, DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), ... | | Authors: | Barski, M.S, Ballandras-Colas, A, Cronin, N.B, Pye, V.E, Cherepanov, P, Maertens, G.N. | | Deposit date: | 2021-06-11 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures.
Nat Commun, 12, 2021
|
|
7OUI
 
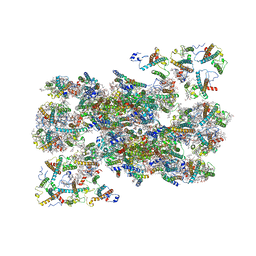 | | Structure of C2S2M2-type Photosystem supercomplex from Arabidopsis thaliana (digitonin-extracted) | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Graca, A.T, Hall, M, Persson, K, Schroder, W.P. | | Deposit date: | 2021-06-11 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | High-resolution model of Arabidopsis Photosystem II reveals the structural consequences of digitonin-extraction.
Sci Rep, 11, 2021
|
|
1C93
 
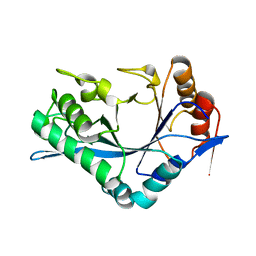 | | Endo-Beta-N-Acetylglucosaminidase H, D130N/E132Q Double Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1CB3
 
 | |
7ONS
 
 | | PARP1 catalytic domain in complex with isoquinolone-based inhibitor (compound 16) | | Descriptor: | 7-[[4-(1,5-dimethylimidazol-2-yl)piperazin-1-yl]methyl]-3-ethyl-1~{H}-quinolin-2-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
6ZCA
 
 | | Structure of the B. subtilis RNA POLYMERASE in complex with HelD (monomer) | | Descriptor: | DNA helicase, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Pei, H.-P, Hilal, T, Huang, Y.-H, Said, N, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-10 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The delta subunit and NTPase HelD institute a two-pronged mechanism for RNA polymerase recycling.
Nat Commun, 11, 2020
|
|
7ONR
 
 | | PARP1 catalytic domain in complex with 8-chloroquinazolinone-based inhibitor (compound 9) | | Descriptor: | 8-chloranyl-2-[3-[4-(1,5-dimethylimidazol-2-yl)piperazin-1-yl]propyl]-3~{H}-quinazolin-4-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
6ZF9
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound SSR4 | | Descriptor: | 1,2-ETHANEDIOL, 3-ethyl-5-phenyl-[1,3]thiazolo[2,3-c][1,2,4]triazole, Bromodomain-containing protein 4 | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound SSR4
To Be Published
|
|
7ONT
 
 | | PARP1 catalytic domain in complex with a selective pyridine carboxamide-based inhibitor (compound 22) | | Descriptor: | 5-[4-[(3-ethyl-2-oxidanylidene-1~{H}-quinolin-7-yl)methyl]piperazin-1-yl]-~{N}-methyl-pyridine-2-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M.J, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
1BTV
 
 | | STRUCTURE OF BET V 1, NMR, 20 STRUCTURES | | Descriptor: | BET V 1 | | Authors: | Osmark, P, Poulsen, F.M, Gajhede, M, Larsen, J.N, Spangfort, M.D. | | Deposit date: | 1997-01-30 | | Release date: | 1997-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy.
Nat.Struct.Biol., 3, 1996
|
|
7OY8
 
 | | Cryo-EM structure of the Rhodospirillum rubrum RC-LH1 complex | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, 2-azanyl-5-[(2~{E},6~{E},8~{E},10~{E},12~{E},14~{E},18~{E},22~{E},26~{E},30~{E},34~{E})-3,7,11,15,19,23,27,31,35,39-decamethyltetraconta-2,6,8,10,12,14,18,22,26,30,34,38-dodecaenyl]-3-methoxy-6-methyl-cyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Qian, P, Croll, T.I, Castro, H.P, Moriarty, N.W, sader, K, Hunter, C.N. | | Deposit date: | 2021-06-23 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of the Rhodospirillum rubrum RC-LH1 complex at 2.5 angstrom.
Biochem.J., 478, 2021
|
|
1BXL
 
 | | STRUCTURE OF BCL-XL/BAK PEPTIDE COMPLEX, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BAK PEPTIDE, BCL-XL | | Authors: | Sattler, M, Liang, H, Nettesheim, D, Meadows, R.P, Harlan, J.E, Eberstadt, M, Yoon, H, Shuker, S.B, Chang, B.S, Minn, A.J, Thompson, C.B, Fesik, S.W. | | Deposit date: | 1996-10-16 | | Release date: | 1997-10-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis.
Science, 275, 1997
|
|
1BJZ
 
 | |
