4LSS
 
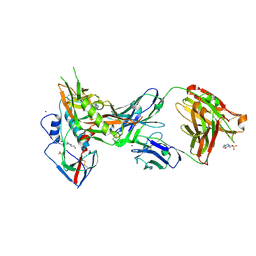 | | Crystal structure of broadly and potently neutralizing antibody VRC01 in complex with HIV-1 clade A strain KER_2018_11 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEAVY CHAIN OF ANTIBODY VRC01, ... | | Authors: | Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Multidonor Analysis Reveals Structural Elements, Genetic Determinants, and Maturation Pathway for HIV-1 Neutralization by VRC01-Class Antibodies.
Immunity, 39, 2013
|
|
5ULG
 
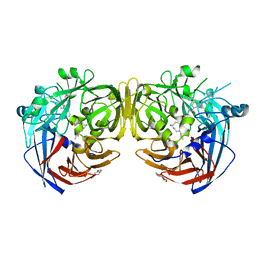 | |
1PPQ
 
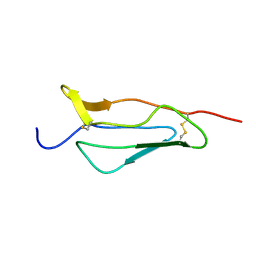 | | NMR structure of 16th module of Immune Adherence Receptor, Cr1 (Cd35) | | Descriptor: | Complement receptor type 1 | | Authors: | O'Leary, J.M, Bromek, K, Black, G.M, Uhrinova, S, Schmitz, C, Krych, M, Atkinson, J.P, Uhrin, D, Barlow, P.N. | | Deposit date: | 2003-06-17 | | Release date: | 2004-05-04 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics of complement control protein (CCP) modules reveals mobility in binding surfaces.
Protein Sci., 13, 2004
|
|
5DEN
 
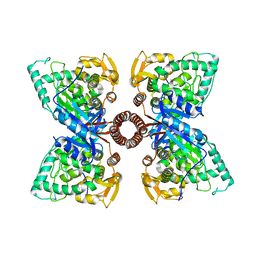 | |
2PR0
 
 | | Crystal structure of Sylvaticin, a new secreted protein from Pythium Sylvaticum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NICKEL (II) ION, sylvaticin | | Authors: | Lascombe, M.B, Prange, T, Retailleau, P. | | Deposit date: | 2007-05-03 | | Release date: | 2008-03-18 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure of sylvaticin, a new alpha-elicitin-like protein from Pythium sylvaticum.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1XON
 
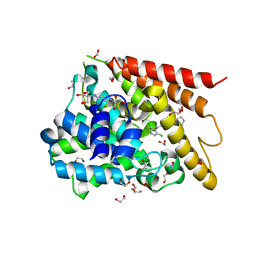 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With Piclamilast | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(CYCLOPENTYLOXY)-N-(3,5-DICHLOROPYRIDIN-4-YL)-4-METHOXYBENZAMIDE, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-06 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
2R5V
 
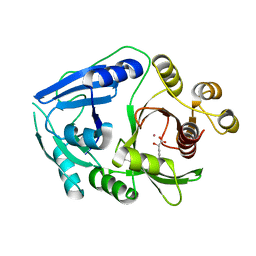 | | Hydroxymandelate Synthase Crystal Structure | | Descriptor: | (2S)-hydroxy(4-hydroxyphenyl)ethanoic acid, COBALT (II) ION, PCZA361.1, ... | | Authors: | Brownlee, J.M, He, P, Moran, G.R, Harrison, D.H.T. | | Deposit date: | 2007-09-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two roads diverged: the structure of hydroxymandelate synthase from Amycolatopsis orientalis in complex with 4-hydroxymandelate.
Biochemistry, 47, 2008
|
|
5UNQ
 
 | |
4BE0
 
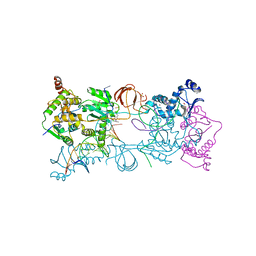 | | PFV intasome with inhibitor XZ-115 | | Descriptor: | 17 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (TRANSFERRED STRAND), 19 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (NON-TRANSFERRED STRAND), 2-(3-chloro-2-fluorobenzyl)-4,5-dihydroxy-1H-isoindole-1,3(2H)-dione, ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2012-10-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Activities, Crystal Structures and Molecular Dynamics of Dihydro-1H-Isoindole Derivatives, Inhibitors of HIV-1 Integrase.
Acs Chem.Biol., 8, 2013
|
|
1G0X
 
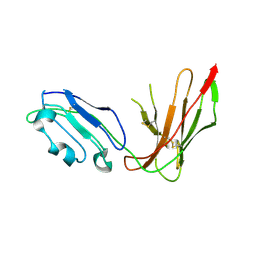 | |
4FZH
 
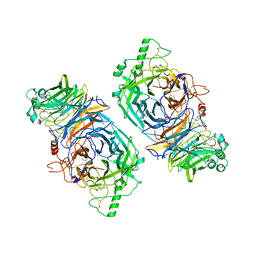 | | Structure of the Ulster Strain Newcastle Disease Virus Hemagglutinin-Neuraminidase Reveals Auto-Inhibitory Interactions Associated with Low Virulence | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-neuraminidase | | Authors: | Yuan, P, Paterson, R.G, Leser, G.P, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2012-07-06 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5008 Å) | | Cite: | Structure of the ulster strain newcastle disease virus hemagglutinin-neuraminidase reveals auto-inhibitory interactions associated with low virulence.
Plos Pathog., 8, 2012
|
|
5PD1
 
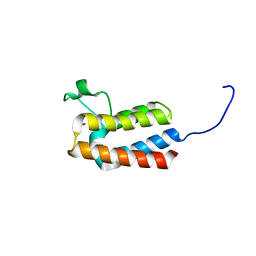 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 57) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PDP
 
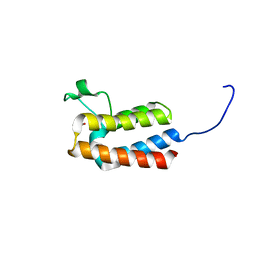 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 82) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PE6
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 99) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5U75
 
 | | The structure of Staphylococcal Enterotoxin-like X (SElX), a Unique Superantigen | | Descriptor: | Enterotoxin-like toxin X, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Ting, Y.T, Young, P.G, Langley, R.J, Baker, H, Fraser, J.D. | | Deposit date: | 2016-12-11 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Staphylococcal enterotoxin-like X (SElX) is a unique superantigen with functional features of two major families of staphylococcal virulence factors.
PLoS Pathog., 13, 2017
|
|
3Q8L
 
 | | Crystal Structure of Human Flap Endonuclease FEN1 (WT) in complex with substrate 5'-flap DNA, SM3+, and K+ | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Classen, S, Chapados, B.R, Arvai, A, Finger, D.L, Guenther, G, Tomlinson, C.G, Thompson, P, Sarker, A.H, Shen, B, Cooper, P.K, Grasby, J.A, Tainer, J.A. | | Deposit date: | 2011-01-06 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | Human Flap Endonuclease Structures, DNA Double-Base Flipping, and a Unified Understanding of the FEN1 Superfamily.
Cell(Cambridge,Mass.), 145, 2011
|
|
3G8I
 
 | | Aleglitazar, a new, potent, and balanced PPAR alpha/gamma agonist for the treatment of type II diabetes | | Descriptor: | (2S)-2-methoxy-3-{4-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]-1-benzothiophen-7-yl}propanoic acid, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor alpha | | Authors: | Benz, J, Bernardeau, A, Binggeli, A, Blum, D, Boehringer, M, Grether, U, Hilpert, H, Kuhn, B, Maerki, H.P, Meyer, M, Puentener, K, Raab, S, Ruf, A, Schlatter, D, Gsell, B, Stihle, M, Mohr, P. | | Deposit date: | 2009-02-12 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aleglitazar, a new, potent, and balanced dual PPARalpha/gamma agonist for the treatment of type II diabetes.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3WUW
 
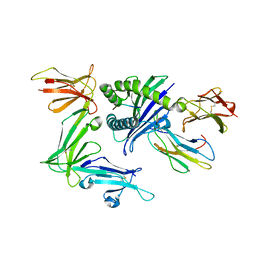 | | KIR3DL1 in complex with HLA-B*57:01.I80T | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2014-05-06 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and structural analysis of KIR3DL1 reveals a lineage-defining allotypic dimorphism that impacts both HLA and peptide sensitivity
J.Immunol., 192, 2014
|
|
2WM5
 
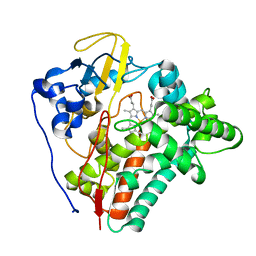 | | X-ray structure of the substrate-free Mycobacterium tuberculosis cytochrome P450 CYP124 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 124 | | Authors: | Johnston, J.B, Kells, P.M, Podust, L.M, Ortiz de Montellano, P.R. | | Deposit date: | 2009-06-30 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and structural characterization of CYP124: a methyl-branched lipid omega-hydroxylase from Mycobacterium tuberculosis.
Proc. Natl. Acad. Sci. U.S.A., 106, 2009
|
|
2X1N
 
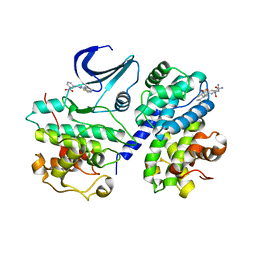 | | Truncation and Optimisation of Peptide Inhibitors of CDK2, Cyclin A Through Structure Guided Design | | Descriptor: | 2-METHYL-N-[(1Z)-3-NITROCYCLOHEXA-2,4-DIEN-1-YLIDENE]-4,5-DIHYDRO[1,3]THIAZOLO[4,5-H]QUINAZOLIN-8-AMINE, ACE-LEU-ASN-PFF-NH2, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Kontopidis, G, Andrews, M.J, McInnes, C, Plater, A, Innes, L, Renachowski, S, Cowan, A, Fischer, P.M, McIntyre, N.A, Griffiths, G, Barnett, A.L, Slawin, A.M.Z, Jackson, W, Thomas, M, Zheleva, D.I, Wang, S, Blake, D.G, Westwood, N.J. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Design, Synthesis, and Evaluation of 2-Methyl- and 2-Amino-N-Aryl-4,5-Dihydrothiazolo[4,5-H]Quinazolin-8-Amines as Ring-Constrained 2-Anilino-4-(Thiazol-5-Yl)Pyrimidine Cyclin-Dependent Kinase Inhibitors.
J.Med.Chem., 53, 2010
|
|
2JES
 
 | | Portal protein (gp6) from bacteriophage SPP1 | | Descriptor: | CALCIUM ION, MERCURY (II) ION, PORTAL PROTEIN, ... | | Authors: | Lebedev, A.A, Krause, M.H, Isidro, A.L, Vagin, A.A, Orlova, E.V, Turner, J, Dodson, E.J, Tavares, P, Antson, A.A. | | Deposit date: | 2007-01-21 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Framework for DNA Translocation Via the Viral Portal Protein
Embo J., 26, 2007
|
|
4RSR
 
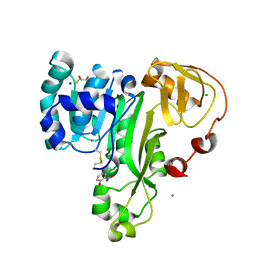 | | ArsM arsenic(III) S-adenosylmethionine methyltransferase with trivalent phenyl arsencial derivative-Roxarsone | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-arsanyl-2-nitrophenol, Arsenic methyltransferase, ... | | Authors: | Packianathan, C, Marapakala, K, Ajees, A.A, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2014-11-10 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A disulfide-bond cascade mechanism for arsenic(III) S-adenosylmethionine methyltransferase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3QE9
 
 | | Crystal structure of human exonuclease 1 Exo1 (D173A) in complex with DNA (complex I) | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*CP*TP*AP*GP*TP*CP*GP*AP*CP*AP*T)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Orans, J, McSweeney, E.A, Iyer, R.R, Hast, M.A, Hellinga, H.W, Modrich, P, Beese, L.S. | | Deposit date: | 2011-01-20 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family.
Cell(Cambridge,Mass.), 145, 2011
|
|
2X8K
 
 | | Crystal Structure of SPP1 Dit (gp 19.1) Protein, a Paradigm of Hub Adsorption Apparatus in Gram-positive Infecting Phages. | | Descriptor: | HYPOTHETICAL PROTEIN 19.1 | | Authors: | Veesler, D, Robin, G, Lichiere, J, Auzat, I, Tavares, P, Bron, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-03-10 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of Bacteriophage Spp1 Distal Tail Protein (Gp 19.1): A Baseplate Hub Paradigm in Gram+ Infecting Phages.
J.Biol.Chem., 285, 2010
|
|
5R4Z
 
 | | XChem fragment screen -- CRYSTAL STRUCTURE OF THE BROMODOMAIN OF THE HUMAN ATAD2 in complex with N13605a | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, SULFATE ION, ... | | Authors: | Talon, R, Krojer, T, Fairhead, M, Sethi, R, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Wright, N, MacLean, E, Zhang, R, Dias, A, Brennan, P.E, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2020-02-28 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | XChem fragment screen
To Be Published
|
|
