2XGK
 
 | | Virus like particle of L172W mutant of Minute Virus of Mice - the immunosuppressive strain | | Descriptor: | COAT PROTEIN VP2 | | Authors: | Plevka, P, Hafenstein, S, Tattersall, P, Cotmore, S, Farr, G, D'Abramo, A, Rossmann, M.G. | | Deposit date: | 2010-06-04 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of a Packaging-Defective Mutant of Minute Virus of Mice Indicates that the Genome is Packaged Via a Pore at a 5-Fold Axis.
J.Virol., 85, 2011
|
|
6ZFW
 
 | | X-ray structure of the soluble N-terminal domain of T. cruzi PEX-14 | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Softley, C.A, Ostertag, M.O, Sattler, M, Popowicz, G.P. | | Deposit date: | 2020-06-18 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Deep learning model predicts water interaction sites on the surface of proteins using limited-resolution data.
Chem.Commun.(Camb.), 56, 2020
|
|
2XNW
 
 | | XPT-PBUX C74U RIBOSWITCH FROM B. SUBTILIS BOUND TO A TRIAZOLO- TRIAZOLE-DIAMINE LIGAND IDENTIFIED BY VIRTUAL SCREENING | | Descriptor: | 3,6-diamino-1,5-dihydro[1,2,4]triazolo[4,3-b][1,2,4]triazol-4-ium, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Daldrop, P, Reyes, F.E, Robinson, D.A, Hammond, C.M, Lilley, D.M.J, Brenk, R. | | Deposit date: | 2010-08-06 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel ligands for a purine riboswitch discovered by RNA-ligand docking.
Chem. Biol., 18, 2011
|
|
6D5C
 
 | |
8RTT
 
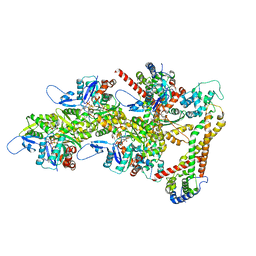 | | Structure of the formin Cdc12 bound to the barbed end of phalloidin-stabilized F-actin. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Molecular mechanism of actin filament elongation by formins.
Science, 384, 2024
|
|
5KE3
 
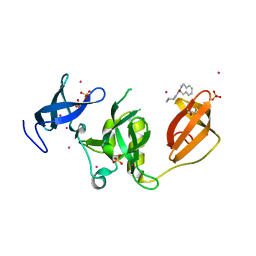 | | Crystal structure of SETDB1 Tudor domain in complex with fragment MRT0181a | | Descriptor: | (S)-N-(furan-2-ylmethyl)-1-(1,2,3,4-tetrahydroisoquinoline-3-carbonyl)piperidine-4-carboxamide, BETA-MERCAPTOETHANOL, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Dong, A, Iqbal, A, Mader, P, Dobrovetsky, E, Ferreira de Freitas, R, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Schapira, M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-09 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain in complex with fragment MRT0181a
to be published
|
|
1W4F
 
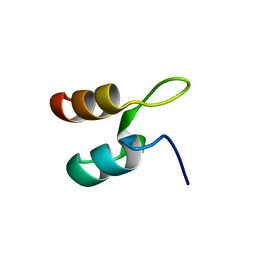 | | Peripheral-subunit from mesophilic, thermophilic and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1VPU
 
 | |
5K2D
 
 | | 1.9A angstrom A2a adenosine receptor structure with MR phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
6Z31
 
 | |
4F8Z
 
 | | Carboxypeptidase T with Boc-Leu | | Descriptor: | CALCIUM ION, Carboxypeptidase T, GLYCEROL, ... | | Authors: | Timofeev, V.I, Kuznetsov, S.A, Akparov, V.K, Kuranova, I.P. | | Deposit date: | 2012-05-18 | | Release date: | 2013-05-22 | | Last modified: | 2015-09-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Three-dimensional structure of carboxypeptidase T from Thermoactinomyces vulgaris in complex with N-BOC-L-leucine.
Biochemistry Mosc., 78, 2013
|
|
1M70
 
 | |
6Z5C
 
 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004070 | | Descriptor: | 7,10-Dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.12,6.017,20]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxylic acid, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004070
To Be Published
|
|
1M9Y
 
 | | X-ray crystal structure of Cyclophilin A/HIV-1 CA N-terminal domain (1-146) M-type H87A,G89A Complex. | | Descriptor: | Cyclophilin A, HIV-1 Capsid | | Authors: | Howard, B.R, Vajdos, F.F, Li, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2002-07-30 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the catalytic mechanism of cyclophilin A
Nat.Struct.Biol., 10, 2003
|
|
6ZJ4
 
 | | apo-Trehalose transferase (apo-TreT) from Thermoproteus uzoniensis | | Descriptor: | THIOCYANATE ION, Trehalose phosphorylase/synthase | | Authors: | Bento, I, Mestrom, L, Marsden, S.R, van der Eijk, H, Laustsen, J.U, Jeffries, C.M, Svergun, D.I, Hagedoorn, P.-H, Hanefeld, U. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anomeric Selectivity of Trehalose Transferase with Rare l-Sugars.
Acs Catalysis, 10, 2020
|
|
2Y5Z
 
 | | Mixed-function P450 MycG in complex with mycinamicin III in C2221 space group | | Descriptor: | BENZAMIDINE, GLYCEROL, MYCINAMICIN III, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-01-19 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
8RTY
 
 | | Structure of the F-actin barbed end bound by Cdc12 and profilin (ring complex) at a resolution of 6.3 Angstrom | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.25 Å) | | Cite: | Molecular mechanism of actin filament elongation by formins.
Science, 384, 2024
|
|
1MP0
 
 | | Binary Complex of Human Glutathione-Dependent Formaldehyde Dehydrogenase with NAD(H) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Sanghani, P.C, Robinson, H, Hurley, T.D, Bosron, W.F. | | Deposit date: | 2002-09-10 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-function relationships in human Class III alcohol dehydrogenase
(formaldehyde dehydrogenase)
Chem.Biol.Interact., 143, 2003
|
|
6DC8
 
 | |
6ZGX
 
 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 1-[2-(2-oxidanylidenepyrrolidin-1-yl)ethyl]-3-phenyl-urea, 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
1MJ3
 
 | | Crystal Structure Analysis of rat enoyl-CoA hydratase in complex with hexadienoyl-CoA | | Descriptor: | ENOYL-COA HYDRATASE, MITOCHONDRIAL, HEXANOYL-COENZYME A | | Authors: | Bell, A.F, Feng, Y, Hofstein, H.A, Parikh, S, Wu, J, Rudolph, M.J, Kisker, C, Tonge, P.J. | | Deposit date: | 2002-08-26 | | Release date: | 2002-09-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stereoselectivity of Enoyl-CoA Hydratase Results from Preferential Activation of
One of Two Bound Substrate Conformers
Chem.Biol., 9, 2002
|
|
1VYQ
 
 | | Novel inhibitors of Plasmodium Falciparum dUTPase provide a platform for anti-malarial drug design | | Descriptor: | 2,3-DEOXY-3-FLUORO-5-O-TRITYLURIDINE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Whittingham, J.L, Leal, I, Kasinathan, G, Nguyen, C, Bell, E, Jones, A.F, Berry, C, Benito, A, Turkenburg, J.P, Dodson, E.J, Ruiz Perez, L.M, Wilkinson, A.J, Johansson, N.G, Brun, R, Gilbert, I.H, Gonzalez Pacanowska, D, Wilson, K.S. | | Deposit date: | 2004-05-05 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dutpase as a Platform for Antimalarial Drug Design: Structural Basis for the Selectivity of a Class of Nucleoside Inhibitors.
Structure, 13, 2005
|
|
4FLP
 
 | | Crystal Structure of the first bromodomain of human BRDT in complex with the inhibitor JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, Bromodomain testis-specific protein, POTASSIUM ION | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Canning, P, Muniz, J, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bradner, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-15 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Small-Molecule Inhibition of BRDT for Male Contraception.
Cell(Cambridge,Mass.), 150, 2012
|
|
2XE7
 
 | | The complete reaction cycle of human phosphoglycerate kinase: The open ternary complex with 3PG and ADP | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, PHOSPHOGLYCERATE KINASE 1 | | Authors: | Cliff, M.J, Baxter, N.J, Blackburn, G.M, Merli, A, Vas, M, Waltho, J.P, Bowler, M.W. | | Deposit date: | 2010-05-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Spring Loaded Release Mechanism Regulates Domain Movement and Catalysis in Phosphoglycerate Kinase.
J.Biol.Chem., 286, 2011
|
|
5K6H
 
 | |
