5F8N
 
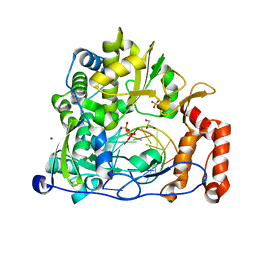 | | Enterovirus 71 Polymerase Elongation Complex (C3S6 Form) | | Descriptor: | GLYCEROL, Genome polyprotein, PYROPHOSPHATE 2-, ... | | Authors: | Shu, B, Gong, P. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.484 Å) | | Cite: | Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3P43
 
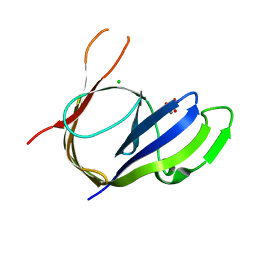 | | Structure and Activities of Archaeal Members of the LigD 3' Phosphoesterase DNA Repair Enzyme Superfamily | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Smith, P, Nair, P.A, Das, U, Shuman, S. | | Deposit date: | 2010-10-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures and activities of archaeal members of the LigD 3'-phosphoesterase DNA repair enzyme superfamily.
Nucleic Acids Res., 39, 2011
|
|
5F8Z
 
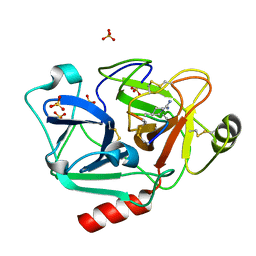 | | The crystal structure of human Plasma Kallikrein in complex with its peptide inhibitor pkalin-1 | | Descriptor: | CYS-PRO-ALA-ARG-PHE-M70-ALA-LEU-PHE-CYS, Plasma kallikrein LIGHT CHAIN, SULFATE ION, ... | | Authors: | Xu, M, Jiang, L, Xu, P, Luo, Z, Andreasen, P, Huang, M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of human Plasma Kallikrein in complex with its peptide inhibitor pkalin-1
To Be Published
|
|
5MKO
 
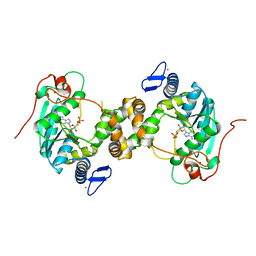 | | [2Fe-2S] cluster containing TtuA in complex with AMP. | | Descriptor: | ADENOSINE MONOPHOSPHATE, FE2/S2 (INORGANIC) CLUSTER, TtuA PH0300, ... | | Authors: | Arragain, S, Bimai, O, Legrand, P, Golinelli-Pimpaneau, B. | | Deposit date: | 2016-12-05 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Nonredox thiolation in tRNA occurring via sulfur activation by a [4Fe-4S] cluster.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MVC
 
 | | Crystal structure of potent human Dihydroorotate Dehydrogenase inhibitors based on hydroxylated azole scaffolds | | Descriptor: | 4-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]-1,2,5-thiadiazole-3-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Andersson, M, Moritzer, A.C, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S, Lolli, M, Friemann, R. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of potent human dihydroorotate dehydrogenase inhibitors based on hydroxylated azole scaffolds.
Eur J Med Chem, 129, 2017
|
|
1BLS
 
 | |
6STJ
 
 | | Selective Affimers Recognize BCL-2 Family Proteins Through Non-Canonical Structural Motifs | | Descriptor: | Cystatin domain-containing protein, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Hobor, F, Miles, J.A, Trinh, C.H, Taylor, J, Tiede, C, Rowell, P.R, Jackson, B, Nadat, F, Kyle, H.F, Wicky, B.I.M, Clarke, J, Tomlinson, D.C, Wilson, A.J, Edwards, T.A. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective Affimers Recognise the BCL-2 Family Proteins BCL-x L and MCL-1 through Noncanonical Structural Motifs*.
Chembiochem, 22, 2021
|
|
3OIH
 
 | | Crystal Structure of the complex of xylanase-alpha-amylase inhibitor Protein (XAIP-I) with trehalose at 1.87 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION, ... | | Authors: | Kumar, M, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-19 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of the complex of xylanase-alpha-amylase inhibitor Protein (XAIP-I) with trehalose at 1.87 A resolution
To be Published
|
|
8X2V
 
 | |
4LMW
 
 | |
7KBT
 
 | | Factor VIII in complex with the anti-C2 domain antibody, G99 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ronayne, E.K, Gish, J, Wilson, C, Peters, S, Spencer, H.T, Spiegel, P.C, Childers, K.C. | | Deposit date: | 2020-10-02 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Structure of Blood Coagulation Factor VIII in Complex With an Anti-C2 Domain Non-Classical, Pathogenic Antibody Inhibitor
Front Immunol, 12, 2021
|
|
6JAI
 
 | | Crystal structure of ABC transporter alpha-glycoside-binding mutant protein D118A in complex with maltose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
5M7I
 
 | | Crystal structure of GH125 1,6-alpha-mannosidase mutant from Clostridium perfringens in complex with 1,6-alpha-mannobiose | | Descriptor: | alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose, exo-alpha-1,6-mannosidase | | Authors: | Males, A, Alonso-Gil, S, Fernandes, P, Williams, S.J, Rovira, C, Davies, G.J. | | Deposit date: | 2016-10-27 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Computational Design of Experiment Unveils the Conformational Reaction Coordinate of GH125 alpha-Mannosidases.
J. Am. Chem. Soc., 139, 2017
|
|
4BOZ
 
 | | Structure of OTUD2 OTU domain in complex with K11-linked di ubiquitin | | Descriptor: | GLYCEROL, UBIQUITIN THIOESTERASE OTU1, UBIQUITIN-C | | Authors: | Mevissen, T.E.T, Hospenthal, M.K, Geurink, P.P, Elliott, P.R, Akutsu, M, Arnaudo, N, Ekkebus, R, Kulathu, Y, Wauer, T, El Oualid, F, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2013-05-22 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Otu Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis.
Cell(Cambridge,Mass.), 154, 2013
|
|
2WB1
 
 | | The complete structure of the archaeal 13-subunit DNA-directed RNA Polymerase | | Descriptor: | DNA-DIRECTED RNA POLYMERASE RPO10 SUBUNIT, DNA-DIRECTED RNA POLYMERASE RPO11 SUBUNIT, DNA-DIRECTED RNA POLYMERASE RPO12 SUBUNIT, ... | | Authors: | Korkhin, Y, Unligil, U.M, Littlefield, O, Nelson, P.J, Stuart, D.I, Sigler, P.B, Bell, S.D, Abrescia, N.G.A. | | Deposit date: | 2009-02-19 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Evolution of Complex RNA Polymerase: The Complete Archaeal RNA Polymerase Structure
Plos Biol., 7, 2009
|
|
1BT5
 
 | | CRYSTAL STRUCTURE OF THE IMIPENEM INHIBITED TEM-1 BETA-LACTAMASE FROM ESCHERICHIA COLI | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, PROTEIN (BETA-LACTAMASE), SULFATE ION | | Authors: | Maveyraud, L, Mourey, L, Pedelacq, J.D, Guillet, V, Kotra, L.K, Mobashery, S, Samama, J.P. | | Deposit date: | 1998-09-02 | | Release date: | 1999-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Clinical Longevity of Carbapenem Antibiotics in the Face of Challenge by the Common Class A Beta-Lactamases from Antibiotic-Resistant Bacteria
J.Am.Chem.Soc., 120, 1998
|
|
8JUA
 
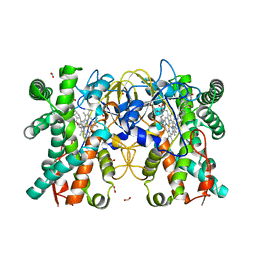 | | Multifunctional cytochrome P450 enzyme IkaD from Streptomyces sp. ZJ306, in complex with epoxyikarugamycin | | Descriptor: | (1Z,3E,5S,7R,8R,10R,11R,12S,13R,15S,16R,17S,19Z,26S)-11-ethyl-2-hydroxy-10-methyl-22,27-diaza-14 oxahexacyclo[24.2.1.05,17.07,16.013,15.08,12]nonacosa-1(2),3,19-triene-21,28,29-trione, Cytochrome P450, FORMIC ACID, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.00001121 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5F8I
 
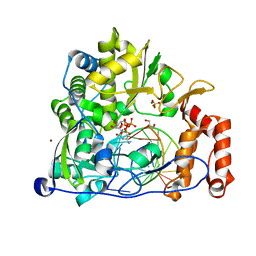 | | Enterovirus 71 Polymerase Elongation Complex (C1S2/3 Form) | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, Genome polyprotein, ... | | Authors: | Shu, B, Gong, P. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1YME
 
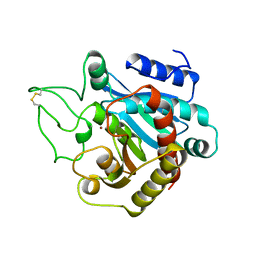 | | STRUCTURE OF CARBOXYPEPTIDASE | | Descriptor: | CARBOXYPEPTIDASE A ALPHA, ZINC ION | | Authors: | Greenblatt, H.M, Tucker, P.A, Shoham, G. | | Deposit date: | 1996-07-15 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Carboxypeptidase A: native, zinc-removed and mercury-replaced forms.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2VEV
 
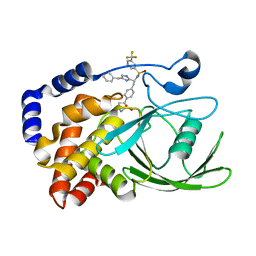 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B IN COMPLEX WITH AN ISOTHIAZOLIDINONE-CONTAINING INHIBITOR | | Descriptor: | MAGNESIUM ION, N-[(1S)-1-(4-benzyl-1H-imidazol-2-yl)-2-{4-[(5S)-1,1-dioxido-3-oxoisothiazolidin-5-yl]phenyl}ethyl]-3-(trifluoromethyl)benzenesulfonamide, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Douty, B, Wayland, B, Ala, P.J, Bower, M.J, Pruitt, J, Bostrom, L, Wei, M, Klabe, R, Gonneville, L, Wynn, R, Burn, T.C, Liu, P.C.C, Combs, A.P, Yue, E.W. | | Deposit date: | 2007-10-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isothiazolidinone Inhibitors of Ptp1B Containing Imidazoles and Imidazolines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4RCR
 
 | | STRUCTURE OF THE REACTION CENTER FROM RHODOBACTER SPHAEROIDES R-26 AND 2.4.1: PROTEIN-COFACTOR (BACTERIOCHLOROPHYLL, BACTERIOPHEOPHYTIN, AND CAROTENOID) INTERACTIONS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Komiya, H, Yeates, T.O, Chirino, A.J, Rees, D.C, Allen, J.P, Feher, G. | | Deposit date: | 1991-09-09 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the reaction center from Rhodobacter sphaeroides R-26 and 2.4.1: protein-cofactor (bacteriochlorophyll, bacteriopheophytin, and carotenoid) interactions.
Proc.Natl.Acad.Sci.USA, 85, 1988
|
|
4RE8
 
 | | Crystal Structure of TR3 LBD in complex with Molecule 5 | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)dodecan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Weijia, W, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
2VIZ
 
 | | Human BACE-1 in complex with N-((1S,2R)-3-(((1S)-2-(cyclohexylamino)- 1-methyl-2-oxoethyl)amino)-2-hydroxy-1-(phenylmethyl)propyl)-3-(2-oxo- 1-pyrrolidinyl)-5-(propyloxy)benzamide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-3-{[(1S)-2-(cyclohexylamino)-1-methyl-2-oxoethyl]amino}-2-hydroxypropyl]-3-(2-oxo-2,3-dihydro-1H-pyrrol-1-yl)-5-propoxybenzamide | | Authors: | Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Faller, A, Hawkins, J, Hussain, I, MacPherson, D, Maile, G, Matico, R, Milner, P, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Riddell, D, Rowland, P, Soleil, V, Smith, K, Stanway, S, Stemp, G, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Ward, J, Wayne, G. | | Deposit date: | 2007-12-06 | | Release date: | 2008-01-29 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bace-1 Inhibitors Part 1: Identification of Novel Hydroxy Ethylamines (Heas).
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6SM2
 
 | | Mutant immunoglobulin light chain causing amyloidosis (Pat-1) | | Descriptor: | Pat-1 | | Authors: | Kazman, P, Vielberg, M.-T, Cendales, M.D.P, Hunziger, L, Weber, B, Hegenbart, U, Zacharias, M, Koehler, R, Schoenland, S, Groll, M, Buchner, J. | | Deposit date: | 2019-08-21 | | Release date: | 2020-03-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fatal amyloid formation in a patient's antibody light chain is caused by a single point mutation.
Elife, 9, 2020
|
|
8OFA
 
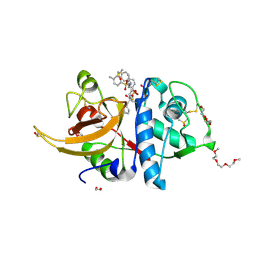 | | Crystal structure of human cathepsin L interacting with tosyl phenylalanyl chloromethyl ketone (TPCK) | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 4-methyl-~{N}-[(2~{S})-4-oxidanyl-3-oxidanylidene-1-phenyl-butan-2-yl]benzenesulfonamide, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Ewert, W, Reinke, P.Y.A, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2023-03-14 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
