7W9M
 
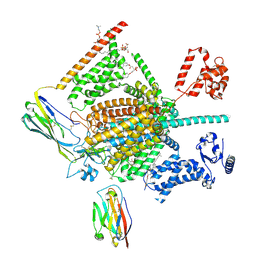 | | Cryo-EM structure of human Nav1.7(E406K) in complex with auxiliary beta subunits, ProTx-II and tetrodotoxin (S6IV pi helix conformer) | | 分子名称: | (1R,5R,6R,7R,9S,11S,12S,13S,14S)-3-amino-14-(hydroxymethyl)-8,10-dioxa-2,4-diazatetracyclo[7.3.1.1~7,11~.0~1,6~]tetradec-3-ene-5,9,12,13,14-pentol (non-preferred name), 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | 登録日 | 2021-12-10 | | 公開日 | 2022-05-25 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
6BND
 
 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, Thr315Ala mutant mono-zinc and phosphoethanolamine complex | | 分子名称: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, POLYETHYLENE GLYCOL (N=34), Phosphoethanolamine transferase, ... | | 著者 | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-11-16 | | 公開日 | 2018-01-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Substrate Recognition by a Colistin Resistance Enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 13, 2018
|
|
6VX8
 
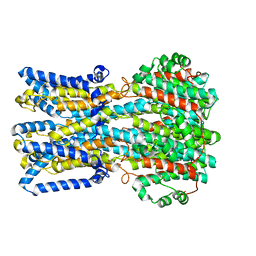 | | bestrophin-2 Ca2+- unbound state 2 (EGTA only) | | 分子名称: | Bestrophin, CHLORIDE ION | | 著者 | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | 登録日 | 2020-02-21 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.33 Å) | | 主引用文献 | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4BNG
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-pentyl- 2-phenoxyphenol | | 分子名称: | 5-PENTYL-2-PHENOXYPHENOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | 著者 | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | 登録日 | 2013-05-15 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
6NJ2
 
 | |
7W9T
 
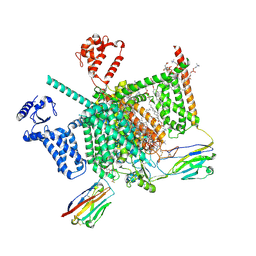 | | Cryo-EM structure of human Nav1.7(E406K) in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin (S6IV alpha helix conformer) | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | 著者 | Yan, N, Huang, G, Liu, D, Wei, P. | | 登録日 | 2021-12-10 | | 公開日 | 2022-05-25 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
3D65
 
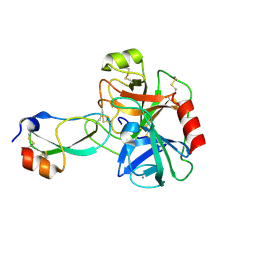 | | Crystal structure of Textilinin-1, a Kunitz-type serine protease inhibitor from the Australian Common Brown snake venom, in complex with trypsin | | 分子名称: | CALCIUM ION, Cationic trypsin, Textilinin | | 著者 | Millers, E.-K.I, Masci, P.P, Lavin, M.F, de Jersey, J, Guddat, L.W. | | 登録日 | 2008-05-19 | | 公開日 | 2009-06-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Crystal structure of Textilinin-1, a Kunitz-type serine protease inhibitor from the Australian Common Brown snake venom, in complex with trypsin
To be Published
|
|
6NKB
 
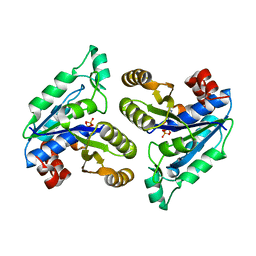 | |
6VL0
 
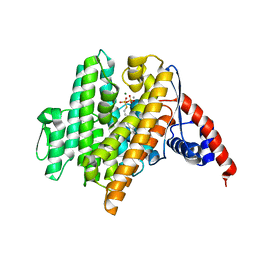 | | Crystal Structure of the N-prenyltransferase DabA in Complex with GSPP and Mn2+ | | 分子名称: | DabA, GERANYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Chekan, J.R, Noel, J.P, Moore, B.S. | | 登録日 | 2020-01-22 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Algal neurotoxin biosynthesis repurposes the terpene cyclase structural fold into anN-prenyltransferase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4H6A
 
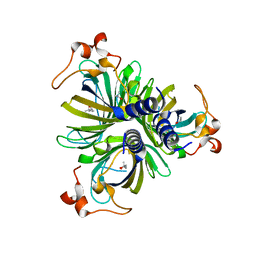 | | Crystal Structure of the Allene Oxide Cyclase 2 from Physcomitrella patens | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Allene oxide cyclase, ... | | 著者 | Neumann, P, Ficner, R. | | 登録日 | 2012-09-19 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structures of Physcomitrella patens AOC1 and AOC2: Insights into the Enzyme Mechanism and Differences in Substrate Specificity.
Plant Physiol., 160, 2012
|
|
6VO8
 
 | | X-ray structure of the Cj1427 in the presence of NADH and GDP-D-glycero-D-mannoheptose, an essential NAD-dependent dehydrogenase from Campylobacter jejuni | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Putative sugar-nucleotide epimerase/dehydratease, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{S},5~{S},6~{S})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | 著者 | Spencer, K.D, Anderson, T.K, Thoden, J.B, Huddleston, J.P, Raushel, F.M, Holden, H.M. | | 登録日 | 2020-01-30 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Analysis of Cj1427, an Essential NAD-Dependent Dehydrogenase for the Biosynthesis of the Heptose Residues in the Capsular Polysaccharides ofCampylobacter jejuni.
Biochemistry, 59, 2020
|
|
6NL4
 
 | |
6NIV
 
 | | Racemic Phenol-Soluble Modulin Alpha 3 Peptide | | 分子名称: | Phenol-soluble modulin PSM-alpha-3 | | 著者 | Yao, Z, Cary, B.P, Bingman, C.A, Wang, C, Kreitler, D.F, Satyshur, K.A, Forest, K.T, Gellman, S.H. | | 登録日 | 2018-12-31 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Use of a Stereochemical Strategy To Probe the Mechanism of Phenol-Soluble Modulin alpha 3 Toxicity.
J.Am.Chem.Soc., 141, 2019
|
|
7WRI
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike receptor-binding domain in complex with mouse ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein, ... | | 著者 | Han, P, Xie, Y, Qi, J. | | 登録日 | 2022-01-26 | | 公開日 | 2022-06-08 | | 最終更新日 | 2022-12-21 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
3D4X
 
 | |
8DNL
 
 | |
1RFN
 
 | | HUMAN COAGULATION FACTOR IXA IN COMPLEX WITH P-AMINO BENZAMIDINE | | 分子名称: | CALCIUM ION, P-AMINO BENZAMIDINE, PROTEIN (COAGULATION FACTOR IX), ... | | 著者 | Hopfner, K.-P, Lang, A, Karcher, A, Sichler, K, Kopetzki, E, Brandstetter, H, Huber, R, Bode, W, Engh, R.A. | | 登録日 | 1999-04-19 | | 公開日 | 1999-09-01 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Coagulation factor IXa: the relaxed conformation of Tyr99 blocks substrate binding.
Structure Fold.Des., 7, 1999
|
|
6BNE
 
 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, phosphate-bound complex | | 分子名称: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-11-16 | | 公開日 | 2018-01-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Substrate recognition by a colistin resistance enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 2018
|
|
3T2V
 
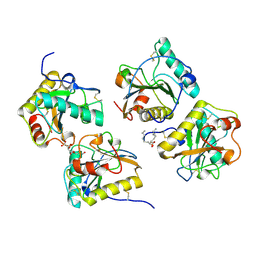 | | Crystal structure of the complex of peptidoglycan recognition protein-short (CPGRP-S) with mycolic acid at 2.5 A resolution | | 分子名称: | (2S,3R)-2-hexyl-3-hydroxynonanoic acid, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2011-07-23 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
6ITP
 
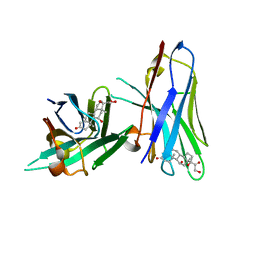 | | Crystal structure of cortisol complexed with its nanobody at pH 3.5 | | 分子名称: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, anti-cortisol camelid antibody | | 著者 | Ding, Y, Ding, L.L, Wang, Z.Y, Zhong, P.Y. | | 登録日 | 2018-11-24 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.572 Å) | | 主引用文献 | Structural insights into the mechanism of single domain VHH antibody binding to cortisol.
Febs Lett., 593, 2019
|
|
6BJ5
 
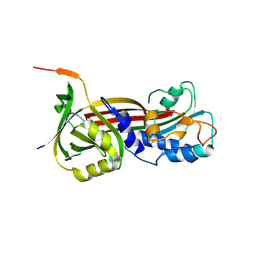 | | Structure of the Clinically used Myxomaviral Serine Protease Inhibitor 1 (SERP-1) | | 分子名称: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Mahon, B.P, Lomelino, C.L, McKenna, R. | | 登録日 | 2017-11-05 | | 公開日 | 2018-11-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of the Clinically used Myxomaviral Serine Protease Inhibitor 1 (SERP-1)
To Be Published
|
|
4KEC
 
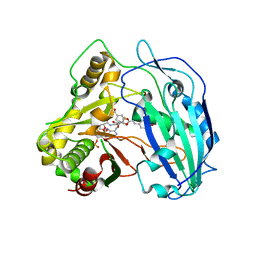 | | SbHCT-complex form | | 分子名称: | (3R,4S,5R)-3,4-dihydroxy-5-{[(2E)-3-(4-hydroxyphenyl)prop-2-enoyl]oxy}cyclohex-1-ene-1-carboxylic acid, COENZYME A, Hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase | | 著者 | Walker, A.M, Hayes, R.P, Youn, B, Vermerris, W, Sattler, S.E, Kang, C. | | 登録日 | 2013-04-25 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other coenzyme a-dependent transferases and synthases.
Plant Physiol., 162, 2013
|
|
8DNA
 
 | |
4HCY
 
 | | Structure of a eukaryotic thiaminase-I bound to the thiamin analogue 3-deazathiamin | | 分子名称: | 2-{4-[(4-amino-2-methylpyrimidin-5-yl)methyl]-3-methylthiophen-2-yl}ethanol, thiaminase-I | | 著者 | Kreinbring, C.A, Hubbard, P.A, Leeper, F.J, Hawksley, D, Petsko, G.A, Ringe, D. | | 登録日 | 2012-10-01 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure of a eukaryotic thiaminase I.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2GEK
 
 | | Crystal Structure of phosphatidylinositol mannosyltransferase (PimA) from Mycobacterium smegmatis in complex with GDP | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHATIDYLINOSITOL MANNOSYLTRANSFERASE (PimA) | | 著者 | Guerin, M.E, Buschiazzo, A, Kordulakova, J, Jackson, M, Alzari, P.M. | | 登録日 | 2006-03-20 | | 公開日 | 2007-04-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria.
J.Biol.Chem., 282, 2007
|
|
