2WEL
 
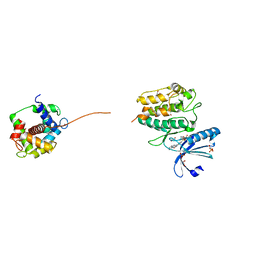 | | Crystal structure of SU6656-bound calcium/calmodulin-dependent protein kinase II delta in complex with calmodulin | | Descriptor: | (3Z)-N,N-DIMETHYL-2-OXO-3-(4,5,6,7-TETRAHYDRO-1H-INDOL-2-YLMETHYLIDENE)-2,3-DIHYDRO-1H-INDOLE-5-SULFONAMIDE, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Pike, A.C.W, Rellos, P, Salah, E, Burgess-Brown, N, Keates, T, Muniz, J, Sethi, R, Roos, A, Filippakopoulos, P, von Delft, F, Edwards, A, Weigelt, J, Arrowsmith, C.H, Bountra, C, Knapp, S. | | Deposit date: | 2009-03-31 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Camkiidelta/Calmodulin Complex Reveals the Molecular Mechanism of Camkii Kinase Activation.
Plos Biol., 8, 2010
|
|
8PT1
 
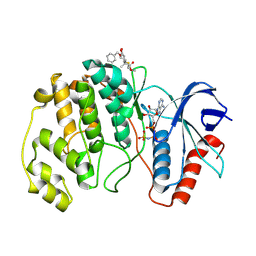 | | ERK2 covelently bound to RU76 cyclohexenone based inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ~{O}1-methyl ~{O}3-(phenylmethyl) (1~{S},3~{R})-1-methyl-4-oxidanylidene-cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
5ORZ
 
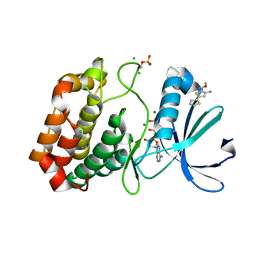 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, CHLORIDE ION, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
5OSE
 
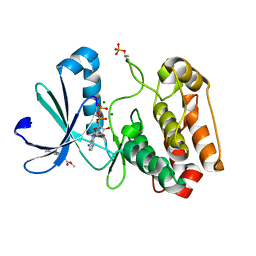 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, MAGNESIUM ION, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-17 | | Release date: | 2017-11-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
8PT3
 
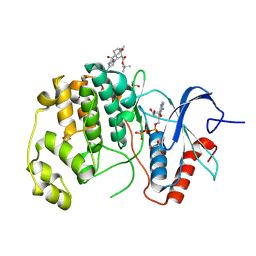 | | ERK2 covelently bound to RU77 cyclohexenone based inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Sok, P, Poti, A, Remenyi, A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
5MOT
 
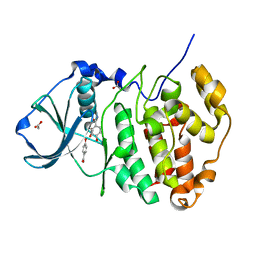 | | Crystal structure of CK2alpha with ZT0627 bound | | Descriptor: | 4-HYDROXYBENZAMIDE, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Georgiou, K, Iegre, J, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2016-12-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
5GJW
 
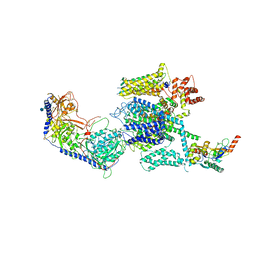 | | Structure of the mammalian voltage-gated calcium channel Cav1.1 complex for ClassII map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, J.P, Yan, Z, Li, Z.Q, Zhou, Q, Yan, N. | | Deposit date: | 2016-07-02 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the voltage-gated calcium channel Cav1.1 at 3.6 angstrom resolution
Nature, 537, 2016
|
|
6JCW
 
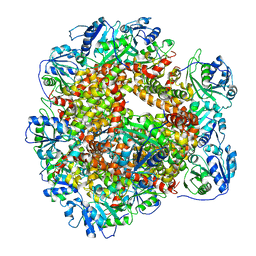 | | Cryo-EM Structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) with Mg2+ at pH8.5 | | Descriptor: | MAGNESIUM ION, ketol-acid reductoisomerase | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
8PVR
 
 | | Cryo-EM structure of horse Nhe9 bound to PI(3,5)P2 | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, Sodium/hydrogen exchanger 9 | | Authors: | Kokane, S, Meier, P, Gulati, A, Delemotte, L, Drew, D. | | Deposit date: | 2023-07-18 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | PIP2 mediated oligomerization of the endosomal sodium/proton exchanger NHE9
To Be Published
|
|
5ORN
 
 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | 3-thiophen-2-yl-4,5-dihydro-1~{H}-pyridazin-6-one, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
5MTL
 
 | | Crystal structure of an amyloidogenic light chain | | Descriptor: | light chain dimer,IGL@ protein,IGL@ protein | | Authors: | Oberti, L, Rognoni, P, Russo, R, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-10 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5ORT
 
 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, CHLORIDE ION, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
1FDO
 
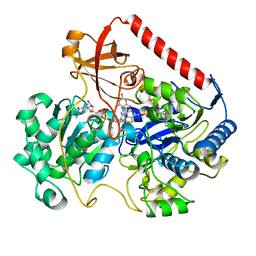 | | OXIDIZED FORM OF FORMATE DEHYDROGENASE H FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FORMATE DEHYDROGENASE H, IRON/SULFUR CLUSTER, ... | | Authors: | Sun, P.D, Boyington, J.C. | | Deposit date: | 1997-01-27 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster.
Science, 275, 1997
|
|
7WSK
 
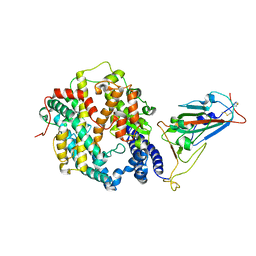 | | Crystal structure of SARS-CoV-2 Omicron spike receptor-binding domain in complex with civet ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Huang, B, Han, P, Qi, J. | | Deposit date: | 2022-01-29 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
5MWJ
 
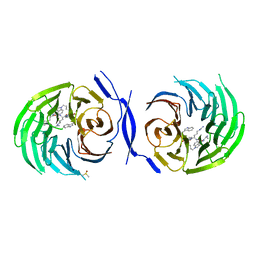 | | Structure Enabled Discovery of a Stapled Peptide Inhibitor to Target the Oncogenic Transcriptional Repressor TLE1 | | Descriptor: | DIMETHYL SULFOXIDE, Transducin-like enhancer protein 1, pepide inhibtor | | Authors: | McGrath, S, Tortorici, M, Vidler, L, Drouin, L, Westwood, I, Gimeson, P, Van Montfort, R, Hoelder, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure-Enabled Discovery of a Stapled Peptide Inhibitor to Target the Oncogenic Transcriptional Repressor TLE1.
Chemistry, 23, 2017
|
|
2BN5
 
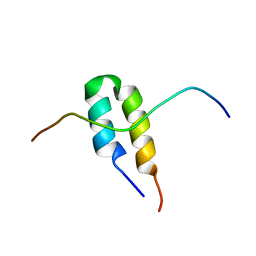 | | P-Element Somatic Inhibitor Protein Complex with U1-70k proline-rich peptide | | Descriptor: | PSI, U1 SMALL NUCLEAR RIBONUCLEOPROTEIN 70 KDA | | Authors: | Ignjatovic, T, Yang, J.-C, Butler, P.J.G, Neuhaus, D, Nagai, K. | | Deposit date: | 2005-03-21 | | Release date: | 2005-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Interaction between P-Element Somatic Inhibitor and U1-70K Essential for the Alternative Splicing of P-Element Transposase.
J.Mol.Biol., 351, 2005
|
|
2BPM
 
 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-630529 | | Descriptor: | (2S)-N-[(3Z)-5-CYCLOPROPYL-3H-PYRAZOL-3-YLIDENE]-2-[4-(2-OXOIMIDAZOLIDIN-1-YL)PHENYL]PROPANAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Cameron, A, Fogliatto, G, Pevarello, P, Brasca, M.G, Orsini, P, Traquandi, G, Longo, A, Nesi, M, Orzi, F, Piutti, C, Sansonna, P, Varasi, M, Vulpetti, A, Roletto, F, Alzani, R, Ciomei, M, Albanese, C, Pastori, W, Marsiglio, A, Pesenti, E, Fiorentini, F, Bischoff, J.R, Mercurio, C. | | Deposit date: | 2005-04-21 | | Release date: | 2005-12-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3-Aminopyrazole Inhibitors of Cdk2-Cyclin a as Antitumor Agents. 2. Lead Optimization
J.Med.Chem., 48, 2005
|
|
3RIX
 
 | | 1.7A resolution structure of a firefly luciferase-Aspulvinone J inhibitor complex | | Descriptor: | (5Z)-4-hydroxy-3-[(2R)-2-(2-hydroxypropan-2-yl)-2,3-dihydro-1-benzofuran-5-yl]-5-{[(2R)-2-(2-hydroxypropan-2-yl)-2,3-dihydro-1-benzofuran-5-yl]methylidene}furan-2(5H)-one, Luciferin 4-monooxygenase | | Authors: | Lovell, S, Battaile, K.P, Lopez, P.C, Auld, D.S, Schultz, P.J, MacArthur, R, Shen, M, Tamayo, G, Inglese, J, Sherman, D.H. | | Deposit date: | 2011-04-14 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Titration-based screening for evaluation of natural product extracts: identification of an aspulvinone family of luciferase inhibitors.
Chem.Biol., 18, 2011
|
|
8A23
 
 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 methyltransferase in complex with TO383 | | Descriptor: | (2R,3R,4S,5R)-2-[4-azanyl-5-(2-quinolin-3-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-5-(hydroxymethyl)oxolane-3,4-diol, 2'-O-methyltransferase nsp16, GLYCEROL, ... | | Authors: | Hanigovsky, M, Krafcikova, P, Klima, M, Boura, E. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 methyltransferase in
complex with TO383
To Be Published
|
|
5GU4
 
 | | rRNA N-glycosylase RTA | | Descriptor: | GLY-PHE-GLY-LEU-PHE-ASP, GLYCEROL, Ricin | | Authors: | Shi, W.W, Tang, Y.S, Sze, S.Y, Zhu, Z.N, Wong, K.B, Shaw, P.C. | | Deposit date: | 2016-08-24 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Ribosome-Inactivating Protein Ricin A Chain in Complex with the C-Terminal Peptide of the Ribosomal Stalk Protein P2
Toxins, 8, 2016
|
|
1PCZ
 
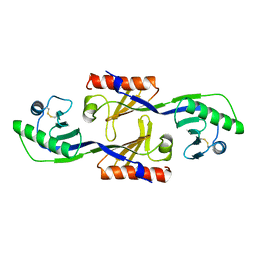 | | STRUCTURE OF TATA-BINDING PROTEIN | | Descriptor: | TATA-BINDING PROTEIN | | Authors: | Dedecker, B.S, Sigler, P.B. | | Deposit date: | 1996-10-04 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a hyperthermophilic archaeal TATA-box binding protein.
J.Mol.Biol., 264, 1996
|
|
8PXB
 
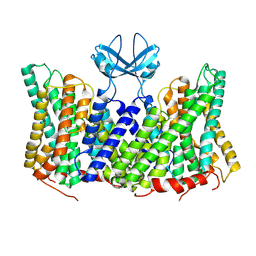 | |
3RMA
 
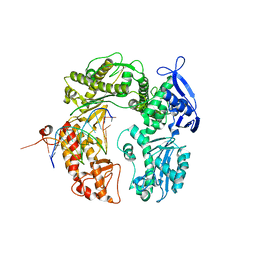 | | Crystal Structure of a replicative DNA polymerase bound to DNA containing Thymine Glycol | | Descriptor: | DNA (5'-D(*CP*GP*AP*(CTG)*GP*AP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*TP*CP*A)-3'), DNA polymerase | | Authors: | Aller, P, Duclos, S, Wallace, S.S, Doublie, S. | | Deposit date: | 2011-04-20 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | A crystallographic study of the role of sequence context in thymine glycol bypass by a replicative DNA polymerase serendipitously sheds light on the exonuclease complex.
J.Mol.Biol., 412, 2011
|
|
5MXX
 
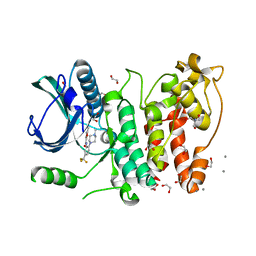 | | Crystal structure of human SR protein kinase 1 (SRPK1) in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-methyl-~{N}-[2-(4-methylpiperazin-1-yl)-5-(trifluoromethyl)phenyl]furan-2-carboxamide, ... | | Authors: | Tallant, C, Redondo, C, Batson, J, Toop, H.D, Babaebi-Jadidib, R, Savitsky, P, Elkins, J.M, Newman, J.A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bates, D.O, Morris, J.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-25 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of Potent, Selective SRPK1 Inhibitors as Potential Topical Therapeutics for Neovascular Eye Disease.
ACS Chem. Biol., 12, 2017
|
|
3J7X
 
 | | Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions | | Descriptor: | Major capsid protein 10A | | Authors: | Guo, F, Liu, Z, Fang, P.A, Zhang, Q, Wright, E.T, Wu, W, Zhang, C, Vago, F, Ren, Y, Jakata, J, Chiu, W, Serwer, P, Jiang, W. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
