1KR3
 
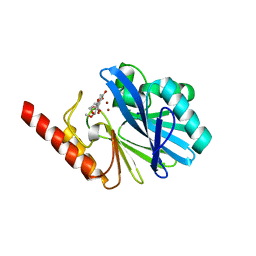 | | Crystal Structure of the Metallo beta-Lactamase from Bacteroides fragilis (CfiA) in Complex with the Tricyclic Inhibitor SB-236050. | | Descriptor: | 7,8-DIHYDROXY-1-METHOXY-3-METHYL-10-OXO-4,10-DIHYDRO-1H,3H-PYRANO[4,3-B]CHROMENE-9-CARBOXYLIC ACID, SODIUM ION, ZINC ION, ... | | Authors: | Payne, D.J, Hueso-Rodrguez, J.A, Boyd, H, Concha, N.O, Janson, C.A, Gilpin, M, Bateson, J.H, Cheever, C, Niconovich, N.L, Pearson, S, Rittenhouse, S, Tew, D, Dez, E, Prez, P, de la Fuente, J, Rees, M, Rivera-Sagredo, A. | | Deposit date: | 2002-01-08 | | Release date: | 2003-01-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a series of tricyclic natural products as potent broad-spectrum inhibitors of metallo-beta-lactamases
ANTIMICROB.AGENTS CHEMOTHER., 46, 2002
|
|
4MC2
 
 | | HIV protease in complex with SA525P | | Descriptor: | (3S)-tetrahydrofuran-3-yl {(2S,3R)-4-[(4R)-4-tert-butyl-7-fluoro-1,1-dioxido-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, Protease | | Authors: | Ganguly, A.K, Alluri, S.S, Wang, C, Caroccia, D, Biswas, D, Kang, E, Zhang, L, Carroll, S.S, Burlein, C, Munshi, V, Orth, P, Strickland, C. | | Deposit date: | 2013-08-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Optimization of Cyclic Sulfonamide based Novel HIV-1 Protease Inhibitors to Pico Molar Affinities guided by X-ray Crystallographic Analysis
Tetrahedron, 2014
|
|
1KKG
 
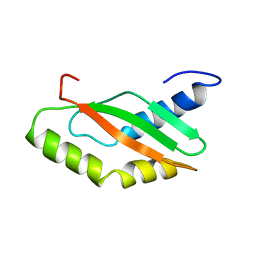 | | NMR Structure of Ribosome-Binding Factor A (RbfA) | | Descriptor: | ribosome-binding factor A | | Authors: | Huang, Y.J, Swapna, G.V.T, Rajan, P.K, Ke, H, Xia, B, Shukla, K, Inouye, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-12-07 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Ribosome-binding Factor A (RbfA), A Cold-shock
Adaptation Protein from Escherichia coli
J.Mol.Biol., 327, 2003
|
|
3JAV
 
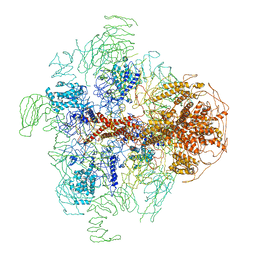 | | Structure of full-length IP3R1 channel in the apo-state determined by single particle cryo-EM | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Fan, G, Baker, M.L, Wang, Z, Baker, M.R, Sinyagovskiy, P.A, Chiu, W, Ludtke, S.J, Serysheva, I.I. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Gating machinery of InsP3R channels revealed by electron cryomicroscopy.
Nature, 527, 2015
|
|
1KSG
 
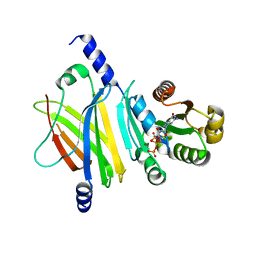 | | Complex of Arl2 and PDE delta, Crystal Form 1 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RETINAL ROD RHODOPSIN-SENSITIVE CGMP 3',5'-CYCLIC PHOSPHODIESTERASE DELTA-SUBUNIT, ... | | Authors: | Hanzal-Bayer, M, Renault, L, Roversi, P, Wittinghofer, A, Hillig, R.C. | | Deposit date: | 2002-01-13 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The complex of Arl2-GTP and PDE delta: from structure to function.
EMBO J., 21, 2002
|
|
6GYX
 
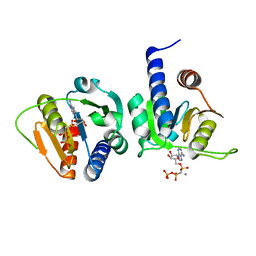 | |
1KLC
 
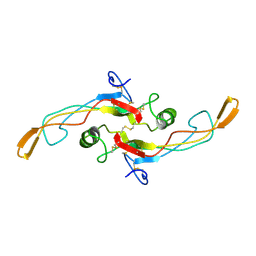 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
4OV8
 
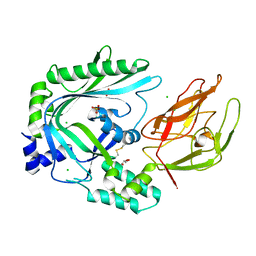 | | Crystal Structure of the TMH1-lock mutant of the mature form of pleurotolysin B | | Descriptor: | CHLORIDE ION, GLYCEROL, Pleurotolysin B, ... | | Authors: | Kondos, S.C, Law, R.H.P, Whisstock, J.C, Dunstone, M.A. | | Deposit date: | 2014-02-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational Changes during Pore Formation by the Perforin-Related Protein Pleurotolysin
Plos Biol., 13, 2015
|
|
1KLD
 
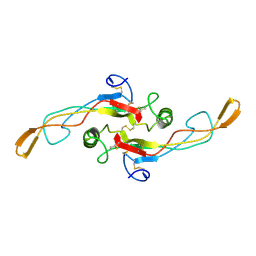 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MODELS 18-33 OF 33 STRUCTURES | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
3IY6
 
 | | Variable domains of the computer generated model (WAM) of Fab E fitted into the cryoEM reconstruction of the virus-Fab E complex | | Descriptor: | fragment from neutralizing antibody E (heavy chain), fragment from neutralizing antibody E (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
1KNK
 
 | | Crystal Structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate Synthase (ispF) from E. coli involved in Mevalonate-Independent Isoprenoid Biosynthesis | | Descriptor: | 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, MANGANESE (II) ION | | Authors: | Richard, S.B, Ferrer, J.L, Bowman, M.E, Lillo, A.M, Tetzlaff, C.N, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-12-18 | | Release date: | 2002-06-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase. An enzyme in the mevalonate-independent isoprenoid biosynthetic pathway.
J.Biol.Chem., 277, 2002
|
|
3J0I
 
 | | Fitting of the phiKZ gp29PR structure into the cryo-EM density map of the phiKZ polysheath | | Descriptor: | PHIKZ029 | | Authors: | Aksyuk, A.A, Fokine, A, Kurochkina, L.P, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2011-08-16 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Structural conservation of the myoviridae phage tail sheath protein fold.
Structure, 19, 2011
|
|
7PDZ
 
 | | Structure of capping protein bound to the barbed end of a cytoplasmic actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Funk, J, Merino, F, Schacks, M, Rottner, K, Raunser, S, Bieling, P. | | Deposit date: | 2021-08-09 | | Release date: | 2021-09-01 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A barbed end interference mechanism reveals how capping protein promotes nucleation in branched actin networks.
Nat Commun, 12, 2021
|
|
7PEL
 
 | | CryoEM structure of simian T-cell lymphotropic virus intasome in complex with PP2A regulatory subunit B56 gamma | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), Isoform 3 of PC4 and SFRS1-interacting protein,Isoform Gamma-2 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, ... | | Authors: | Barski, M, Pye, V.E, Nans, A, Cherepanov, P, Maertens, G.N. | | Deposit date: | 2021-08-10 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of the deltaretroviral intasome in complex with the PP2A regulatory subunit B56gamma.
Nat Commun, 11, 2020
|
|
6ZUL
 
 | | Crystal structure of dimethylated RSL in complex with cucurbit[7]uril and zinc | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, SODIUM ION, ... | | Authors: | Guagnini, F, Engilberge, S, Flood, R.J, Ramberg, K.O, Crowley, P.B. | | Deposit date: | 2020-07-23 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Metal-Mediated Protein-Cucurbituril Crystalline Architectures
Cryst.Growth Des., 2020
|
|
3IZX
 
 | | 3.1 Angstrom cryoEM structure of cytoplasmic polyhedrosis virus | | Descriptor: | Capsid protein VP1, Structural protein VP3, Viral structural protein 5 | | Authors: | Yu, X, Ge, P, Jiang, J, Atanasov, I, Zhou, Z.H. | | Deposit date: | 2011-01-15 | | Release date: | 2011-06-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Atomic Model of CPV Reveals the Mechanism Used by This Single-Shelled Virus to Economically Carry Out Functions Conserved in Multishelled Reoviruses.
Structure, 19, 2011
|
|
1KW1
 
 | | Crystal Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase with bound L-gulonate 6-phosphate | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, L-GULURONIC ACID 6-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E, Yew, W.S, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | Deposit date: | 2002-01-28 | | Release date: | 2002-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Homologous (beta/alpha)8-barrel enzymes that catalyze unrelated reactions: orotidine 5'-monophosphate decarboxylase and 3-keto-L-gulonate 6-phosphate decarboxylase.
Biochemistry, 41, 2002
|
|
3IY5
 
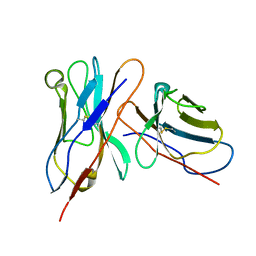 | | Variable domains of the mouse Fab (1AIF) fitted into the cryoEM reconstruction of the virus-Fab 16 complex | | Descriptor: | antibody fragment IGG2A (heavy chain), antibody fragment IGG2A (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
4R5A
 
 | |
3IY3
 
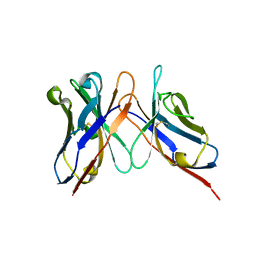 | | Variable domains of the computer generated model (WAM) of Fab 8 fitted into the cryoEM reconstruction of the virus-Fab 8 complex | | Descriptor: | antibody fragment from neutralizing antibody 8 (heavy chain), antibody fragment from neutralizing antibody 8 (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (11.1 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IYK
 
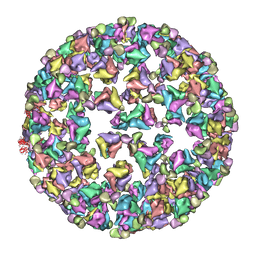 | | Bluetongue virus structure reveals a sialic acid binding domain, amphipathic helices and a central coiled coil in the outer capsid proteins | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, VP2, VP5 | | Authors: | Zhang, X, Boyce, M, Bhattacharya, B, Zhang, X, Schein, S, Roy, P, Zhou, Z.H. | | Deposit date: | 2010-01-25 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Bluetongue virus coat protein VP2 contains sialic acid-binding domains, and VP5 resembles enveloped virus fusion proteins.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4LW6
 
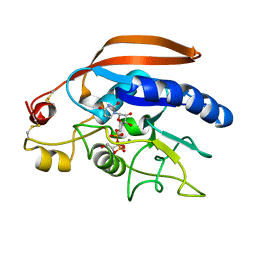 | | Crystal structure of catalytic domain of Drosophila beta1,4galactosyltransferase 7 complex with xylobiose | | Descriptor: | 2-AMINO-1,3-PROPANEDIOL, Beta-4-galactosyltransferase 7, MANGANESE (II) ION, ... | | Authors: | Qasba, P.K, Ramakrishnan, B. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of beta-1,4-Galactosyltransferase 7 Enzyme Reveal Conformational Changes and Substrate Binding.
J.Biol.Chem., 288, 2013
|
|
7A96
 
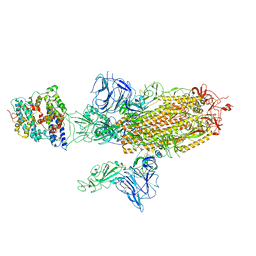 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound and 1 RBD Erect in Anticlockwise Direction | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
4R5K
 
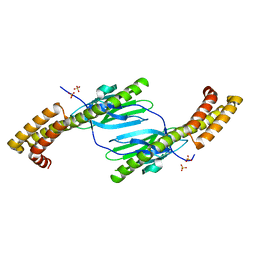 | | Crystal structure of the DnaK C-terminus (Dnak-SBD-B) | | Descriptor: | CALCIUM ION, Chaperone protein DnaK, SULFATE ION | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7469 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
3J0H
 
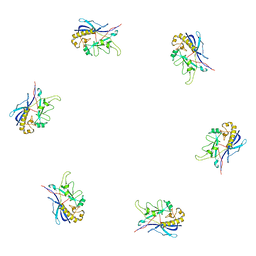 | | Fitting of the bacteriophage phiKZ gp29PR structure into the cryo-EM density map of the phiKZ extended tail sheath | | Descriptor: | PHIKZ029 | | Authors: | Aksyuk, A.A, Fokine, A, Kurochkina, L.P, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2011-08-16 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structural conservation of the myoviridae phage tail sheath protein fold.
Structure, 19, 2011
|
|
