2GM9
 
 | | Structure of rabbit muscle glycogen phosphorylase in complex with thienopyrrole | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 2-CHLORO-N-[(3R)-2-OXO-1,2,3,4-TETRAHYDROQUINOLIN-3-YL]-6H-THIENO[2,3-B]PYRROLE-5-CARBOXAMIDE, Glycogen phosphorylase, ... | | Authors: | Otterbein, L.R, Pannifer, A.D, Tucker, J, Breed, J, Oikonomakos, N.G, Minshull, C, Rowsell, S, Pauptit, R.A. | | Deposit date: | 2006-04-06 | | Release date: | 2007-02-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel thienopyrrole glycogen phosphorylase inhibitors: synthesis, in vitro SAR and crystallographic studies.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4R0E
 
 | |
8B7V
 
 | | Automated simulation-based refinement of maltoporin into a cryo-EM density | | Descriptor: | Maltoporin | | Authors: | Yvonnesdotter, L, Rovsnik, U, Blau, C, Lycksell, M, Howard, R.J, Lindahl, E. | | Deposit date: | 2022-10-03 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Automated simulation-based membrane protein refinement into cryo-EM data.
Biophys.J., 122, 2023
|
|
2GJ4
 
 | | Structure of rabbit muscle glycogen phosphorylase in complex with ligand | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 2-CHLORO-N-[(1R,2R)-1-HYDROXY-2,3-DIHYDRO-1H-INDEN-2-YL]-6H-THIENO[2,3-B]PYRROLE-5-CARBOXAMIDE, Glycogen phosphorylase, ... | | Authors: | Otterbein, L.R, Pannifer, A.D, Tucker, J, Breed, J, Oikonomakos, N.G, Rowsell, S, Pauptit, R.A, Claire, M. | | Deposit date: | 2006-03-30 | | Release date: | 2007-02-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel thienopyrrole glycogen phosphorylase inhibitors: synthesis, in vitro SAR and crystallographic studies.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2IJD
 
 | | Crystal Structure of the Poliovirus Precursor Protein 3CD | | Descriptor: | Picornain 3C, RNA-directed RNA polymerase, SULFATE ION, ... | | Authors: | Marcotte, L.L, Gohara, D.W, Filman, D.J, Hogle, J.M. | | Deposit date: | 2006-09-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of poliovirus 3CD: virally-encoded protease and precursor to the RNA-dependent RNA polymerase.
J.Virol., 81, 2007
|
|
2IJF
 
 | |
1K8U
 
 | |
1K96
 
 | | CRYSTAL STRUCTURE OF CALCIUM BOUND HUMAN S100A6 | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, S100A6 | | Authors: | Otterbein, L.R, Dominguez, R. | | Deposit date: | 2001-10-26 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structures of S100A6 in the Ca(2+)-free and Ca(2+)-bound states: the calcium sensor mechanism of S100 proteins revealed at atomic resolution.
Structure, 10, 2002
|
|
1K9K
 
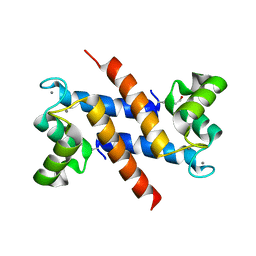 | | CRYSTAL STRUCTURE OF CALCIUM BOUND HUMAN S100A6 | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, S100A6 | | Authors: | Otterbein, L.R, Dominguez, R. | | Deposit date: | 2001-10-29 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structures of S100A6 in the Ca(2+)-free and Ca(2+)-bound states: the calcium sensor mechanism of S100 proteins revealed at atomic resolution.
Structure, 10, 2002
|
|
1K9P
 
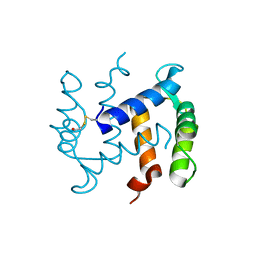 | |
2H8H
 
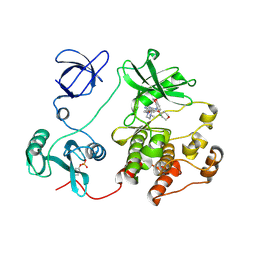 | | Src kinase in complex with a quinazoline inhibitor | | Descriptor: | N-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-7-[2-(4-METHYLPIPERAZIN-1-YL)ETHOXY]-5-(TETRAHYDRO-2H-PYRAN-4-YLOXY)QUINAZOLIN-4-AMINE, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Otterbein, L.R, Norman, R, Pauptit, R.A, Rowsell, S, Breed, J. | | Deposit date: | 2006-06-07 | | Release date: | 2006-11-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | N-(5-chloro-1,3-benzodioxol-4-yl)-7-[2-(4-methylpiperazin-1-yl)ethoxy]-5- (tetrahydro-2H-pyran-4-yloxy)quinazolin-4-amine, a novel, highly selective, orally available, dual-specific c-Src/Abl kinase inhibitor.
J.Med.Chem., 49, 2006
|
|
1KW2
 
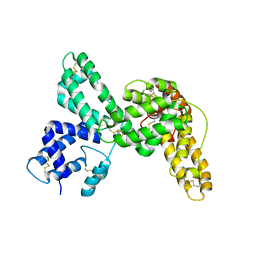 | |
1KXP
 
 | |
4AA7
 
 | | E.coli GlmU in complex with an antibacterial inhibitor | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, N-(2,4-dimethoxy-5-{[(2R)-2-methyl-2,3-dihydro-1H-indol-1-yl]sulfonyl}phenyl)acetamide, SULFATE ION | | Authors: | Otterbein, L, Breed, J, Ogg, D.J. | | Deposit date: | 2011-11-30 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitors of Acetyltransferase Domain of N-Acetylglucosamine-1-Phosphate-Uridyltransferase/ Glucosamine-1-Phosphate-Acetyltransferase (Glmu). Part 1: Hit to Lead Evaluation of a Novel Arylsulfonamide Series.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4AAW
 
 | | S.pneumoniae GlmU in complex with an antibacterial inhibitor | | Descriptor: | 4-{[1-(2-{[({5-[(3-carboxypropanoyl)amino]-2,4-dimethoxyphenyl}sulfonyl)amino]methyl}phenyl)piperidin-4-yl]methoxy}-4-oxobutanoic acid, BIFUNCTIONAL PROTEIN GLMU, SULFATE ION | | Authors: | Otterbein, L, Breed, J, Ogg, D.J. | | Deposit date: | 2011-12-05 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of Acetyltransferase Domain of N-Acetylglucosamine-1-Phosphate-Uridyltransferase/ Glucosamine-1-Phosphate-Acetyltransferase (Glmu). Part 1: Hit to Lead Evaluation of a Novel Arylsulfonamide Series.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4AC3
 
 | | S.pneumoniae GlmU in complex with an antibacterial inhibitor | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, N-{2,4-DIMETHOXY-5-[(2-PIPERIDIN-1-YLBENZYL)sulfamoyl]phenyl}acetamide, SULFATE ION | | Authors: | Otterbein, L, Breed, J, Ogg, D.J. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of Acetyltransferase Domain of N-Acetylglucosamine-1-Phosphate-Uridyltransferase/ Glucosamine-1-Phosphate-Acetyltransferase (Glmu). Part 1: Hit to Lead Evaluation of a Novel Arylsulfonamide Series.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1J6Z
 
 | | UNCOMPLEXED ACTIN | | Descriptor: | ACTIN ALPHA 1, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Otterbein, L.R, Graceffa, P, Dominguez, R. | | Deposit date: | 2001-05-15 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The crystal structure of uncomplexed actin in the ADP state.
Science, 293, 2001
|
|
1ZEA
 
 | | Structure of the anti-cholera toxin antibody Fab fragment TE33 in complex with a D-peptide | | Descriptor: | CITRIC ACID, monoclonal anti-cholera toxin IGG1 KAPPA antibody, H chain, ... | | Authors: | Scheerer, P, Krauss, N, Wessner, H, Scholz, C, Otte, L, Seifert, M, Kramer, A, Schneider-Mergener, J, Hoehne, W. | | Deposit date: | 2005-04-18 | | Release date: | 2006-04-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of an anti-cholera toxin antibody Fab in complex with an epitope-derived D-peptide: a case of polyspecific recognition.
J.Mol.Recognit., 20, 2007
|
|
1YWI
 
 | | Structure of the FBP11WW1 domain complexed to the peptide APPTPPPLPP | | Descriptor: | Formin, Formin-binding protein 3 | | Authors: | Pires, J.R, Parthier, C, Aido-Machado, R, Wiedemann, U, Otte, L, Boehm, G, Rudolph, R, Oschkinat, H. | | Deposit date: | 2005-02-18 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for APPTPPPLPP peptide recognition by the FBP11WW1 domain.
J.Mol.Biol., 348, 2005
|
|
1YWJ
 
 | | Structure of the FBP11WW1 domain | | Descriptor: | Formin-binding protein 3 | | Authors: | Pires, J.R, Parthier, C, Aido-Machado, R, Wiedemann, U, Otte, L, Boehm, G, Rudolph, R, Oschkinat, H. | | Deposit date: | 2005-02-18 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for APPTPPPLPP peptide recognition by the FBP11WW1 domain.
J.Mol.Biol., 348, 2005
|
|
4UCT
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-amino-6-methyl-5-(propan-2-yloxy)-3H-[1,2,4]triazolo[1,5-a]pyrimidin-8-ium, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCU
 
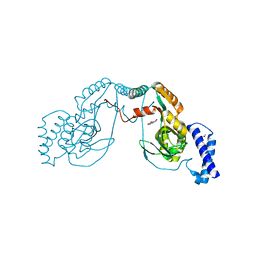 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-hydroxyquinoline-2-carboxylic acid, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCS
 
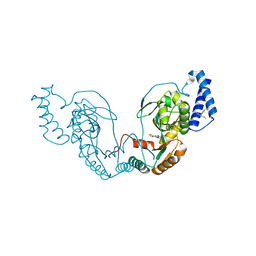 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 5-amino-3-(furan-2-yl)-1H-1,2,4-triazole-1-carboxamide, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCO
 
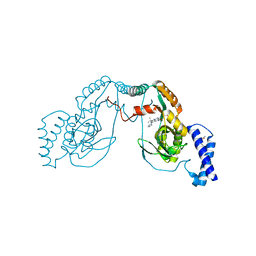 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 7-amino-2-tert-butyl-4-(1H-pyrrol-2-yl)pyrido[2,3-d]pyrimidine-6-carboxamide, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCV
 
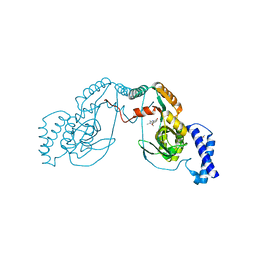 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-methoxy-2,3-dimethylquinoxalin-5-ol, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
