4TNT
 
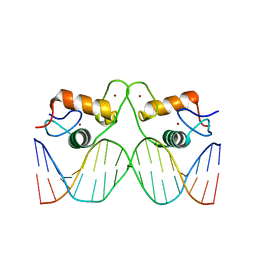 | | Structure of the human mineralocorticoid receptor in complex with DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), Mineralocorticoid receptor, ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2014-06-04 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3938 Å) | | Cite: | Crystal structure of the mineralocorticoid receptor DNA binding domain in complex with DNA.
Plos One, 9, 2014
|
|
8GDR
 
 | |
6X6E
 
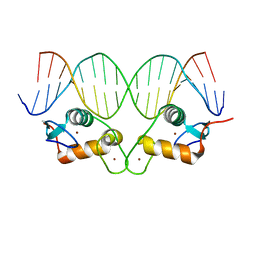 | |
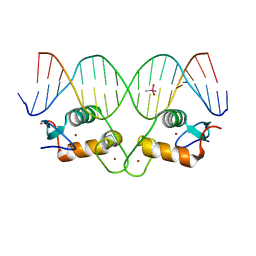
6X6D
 
 | |
6CFN
 
 | |
6C1Z
 
 | |
7TT8
 
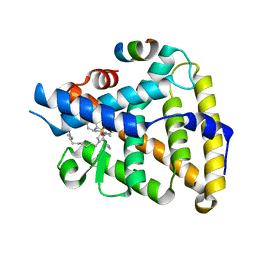 | | Human LRH-1 LBD bound to agonist 6N-10CA and fragment of Tif2 coactivator | | Descriptor: | 10-[(3aR,6S,6aR)-3-phenyl-3a-(1-phenylethenyl)-6-(sulfamoylamino)-1,3a,4,5,6,6a-hexahydropentalen-2-yl]decanoic acid (non-preferred name), Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Cato, M.L, Ortlund, E.A. | | Deposit date: | 2022-01-31 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Differential Modulation of Nuclear Receptor LRH-1 through Targeting Buried and Surface Regions of the Binding Pocket.
J.Med.Chem., 65, 2022
|
|
4DOR
 
 | |
4DOS
 
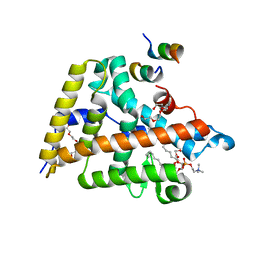 | | Human Nuclear Receptor Liver Receptor Homologue-1, LRH-1, Bound to DLPC and a Fragment of TIF-2 | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2, ... | | Authors: | Musille, P.M, Ortlund, E.A. | | Deposit date: | 2012-02-10 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antidiabetic phospholipid-nuclear receptor complex reveals the mechanism for phospholipid-driven gene regulation.
Nat.Struct.Mol.Biol., 19, 2012
|
|
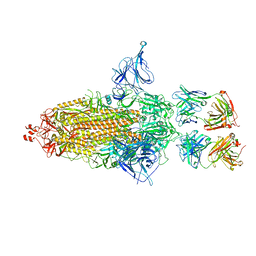
8EYH
 
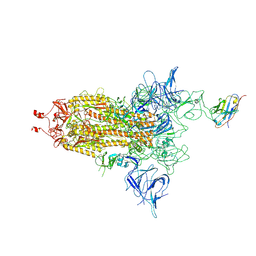 | | SARS-CoV-2 spike protein bound with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody, ... | | Authors: | Laughlin, Z.T, Patel, A, Ortlund, E.A. | | Deposit date: | 2022-10-27 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | SARS-CoV-2 spike protein bound with nanobody
To Be Published
|
|
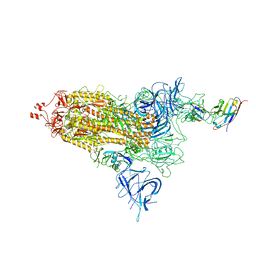
8EYG
 
 | |
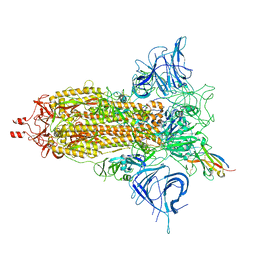
8F4P
 
 | |
8F8M
 
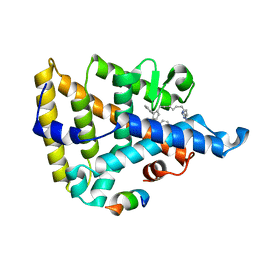 | | LRH-1 bound to small molecule Tet and fragment of coactivator Tif2 | | Descriptor: | (1~{R},3~{a}~{R},6~{a}~{R})-4-phenyl-3~{a}-(1-phenylethenyl)-5-[9-(1~{H}-1,2,3,4-tetrazol-5-yl)nonyl]-2,3,6,6~{a}-tetrahydro-1~{H}-pentalen-1-ol, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Cato, M.L, Ortlund, E.A. | | Deposit date: | 2022-11-22 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Isosteric improvements to liver receptor homolog-1 small molecule modulators
To Be Published
|
|
5L0M
 
 | | hLRH-1 DNA Binding Domain - 12bp Oct4 promoter complex | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*CP*CP*TP*TP*GP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*CP*AP*AP*GP*GP*CP*TP*AP*G)-3'), GLYCEROL, ... | | Authors: | Tuntland, M.L, Ortlund, E.A. | | Deposit date: | 2016-07-27 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | A Structural Investigation into Oct4 Regulation by Orphan Nuclear Receptors, Germ Cell Nuclear Factor (GCNF), and Liver Receptor Homolog-1 (LRH-1).
J. Mol. Biol., 428, 2016
|
|
5KRB
 
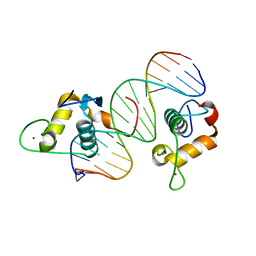 | | GCNF DNA Binding Domain - Oct4 DR0 Complex | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*GP*TP*CP*AP*AP*GP*GP*CP*TP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*AP*GP*CP*CP*TP*TP*GP*AP*CP*CP*TP*CP*T)-3'), Nuclear receptor subfamily 6 group A member 1, ... | | Authors: | Weikum, E.R, Ortlund, E.A. | | Deposit date: | 2016-07-07 | | Release date: | 2016-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | A Structural Investigation into Oct4 Regulation by Orphan Nuclear Receptors, Germ Cell Nuclear Factor (GCNF), and Liver Receptor Homolog-1 (LRH-1).
J. Mol. Biol., 428, 2016
|
|
5L11
 
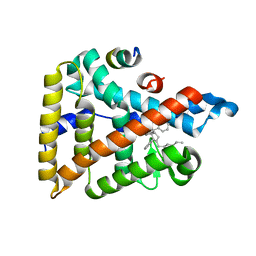 | | Human Liver Receptor Homologue-1 (LRH-1) Bound to RJW100 and a Fragment of TIF-2 | | Descriptor: | (1R,3aR,6aR)-5-hexyl-4-phenyl-3a-(1-phenylethenyl)-1,2,3,3a,6,6a-hexahydropentalen-1-ol, Nuclear receptor subfamily 5 group A member 2, Tif2 | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2016-07-28 | | Release date: | 2016-10-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Crystal Structures of the Nuclear Receptor, Liver Receptor Homolog 1, Bound to Synthetic Agonists.
J. Biol. Chem., 291, 2016
|
|
6NWL
 
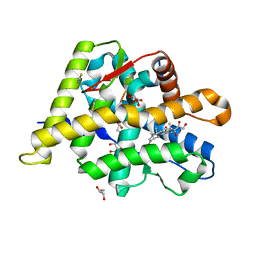 | | Structure of the Ancestral Glucocorticoid Receptor 2 ligand binding domain in complex with hydrocortisone and PGC1a coregulator fragment | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Liu, X, Ortlund, E.A. | | Deposit date: | 2019-02-06 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.595 Å) | | Cite: | First High-Resolution Crystal Structures of the Glucocorticoid Receptor Ligand-Binding Domain-Peroxisome Proliferator-ActivatedgammaCoactivator 1-alphaComplex with Endogenous and Synthetic Glucocorticoids.
Mol.Pharmacol., 96, 2019
|
|
6NWK
 
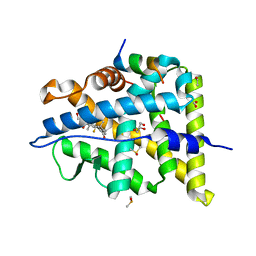 | |
5VA7
 
 | | Glucocorticoid Receptor DNA Binding Domain - IL11 AP-1 recognition element Complex | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*GP*AP*GP*TP*CP*AP*GP*GP*AP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*CP*CP*TP*GP*AP*CP*TP*CP*AP*CP*CP*CP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Weikum, E.R, Ortlund, E.A. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Tethering not required: the glucocorticoid receptor binds directly to activator protein-1 recognition motifs to repress inflammatory genes.
Nucleic Acids Res., 45, 2017
|
|
5UNJ
 
 | | Structure of Human Liver Receptor Homolog 1 in complex with PGC1a and RJW100 | | Descriptor: | (1R,3aR,6aR)-5-hexyl-4-phenyl-3a-(1-phenylethenyl)-1,2,3,3a,6,6a-hexahydropentalen-1-ol, Nuclear receptor subfamily 5 group A member 2, Peroxisome proliferator-activated gamma coactivator 1-alpha | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2017-01-31 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.959 Å) | | Cite: | Structure and Dynamics of the Liver Receptor Homolog 1-PGC1 alpha Complex.
Mol. Pharmacol., 92, 2017
|
|
5VA0
 
 | |
5UFS
 
 | |
6OQX
 
 | | Human Liver Receptor Homolog-1 bound to the agonist 5N and a fragment of the Tif2 coregulator | | Descriptor: | (8beta,11alpha,12alpha)-8-(1-phenylethenyl)-1,6:7,14-dicycloprosta-1,3,5,7(14)-tetraen-11-yl sulfamate, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2019-04-29 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Development of the First Low Nanomolar Liver Receptor Homolog-1 Agonist through Structure-guided Design.
J.Med.Chem., 62, 2019
|
|
6OR1
 
 | | Human LRH-1 bound to the agonist 2N and a fragment of the Tif2 coregulator | | Descriptor: | N-[(1S,3aR,6aR)-5-hexyl-4-phenyl-3a-(1-phenylethenyl)-1,2,3,3a,6,6a-hexahydropentalen-1-yl]acetamide, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2019-04-29 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Development of the First Low Nanomolar Liver Receptor Homolog-1 Agonist through Structure-guided Design.
J.Med.Chem., 62, 2019
|
|
6OQY
 
 | | Human LRH-1 bound to the agonist 6N and a fragment of the Tif2 coregulator | | Descriptor: | N-[(1S,3aR,6aR)-5-hexyl-4-phenyl-3a-(1-phenylethenyl)-1,2,3,3a,6,6a-hexahydropentalen-1-yl]sulfuric diamide, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2019-04-29 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Development of the First Low Nanomolar Liver Receptor Homolog-1 Agonist through Structure-guided Design.
J.Med.Chem., 62, 2019
|
|
