5J3Z
 
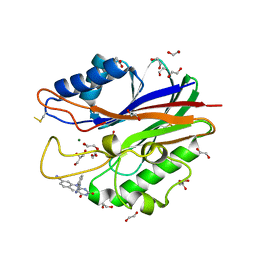 | | Crystal structure of m2hTDP2-CAT in complex with a small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dioxo-10-[3-(1H-tetrazol-5-yl)phenyl]-2,3,4,10-tetrahydropyrimido[4,5-b]quinoline-8-carbonitrile, ACETATE ION, ... | | Authors: | Hornyak, P, Pearl, L.H, Caldecott, K.W, Oliver, A.W. | | Deposit date: | 2016-03-31 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2.
Biochem.J., 473, 2016
|
|
5ECG
 
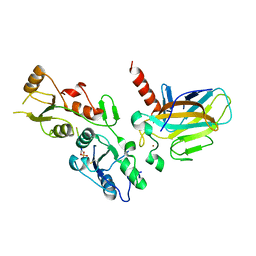 | | Crystal structure of the BRCT domains of 53BP1 in complex with p53 and H2AX-pSer139 (gammaH2AX) | | Descriptor: | Cellular tumor antigen p53, SEP-GLN-GLU-TYR, Tumor suppressor p53-binding protein 1, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ATM Localization and Heterochromatin Repair Depend on Direct Interaction of the 53BP1-BRCT2 Domain with gamma H2AX.
Cell Rep, 13, 2015
|
|
5J3P
 
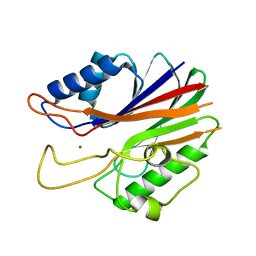 | | Crystal structure of the catalytic domain of human tyrosyl DNA phosphodiesterase 2 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Tyrosyl-DNA phosphodiesterase 2 | | Authors: | Hornyak, P, Pearl, L.H, Caldecott, K.W, Oliver, A.W. | | Deposit date: | 2016-03-31 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2.
Biochem.J., 473, 2016
|
|
6HM5
 
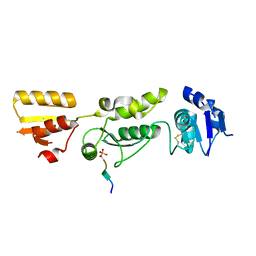 | | Crystal structure of TOPBP1 BRCT0,1,2 in complex with a RAD9 phosphopeptide | | Descriptor: | Cell cycle checkpoint control protein RAD9A, DNA topoisomerase II binding protein 1 | | Authors: | Day, M, Rappas, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2018-09-12 | | Release date: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (2.330038 Å) | | Cite: | BRCT domains of the DNA damage checkpoint proteins TOPBP1/Rad4 display distinct specificities for phosphopeptide ligands.
Elife, 7, 2018
|
|
5MG8
 
 | | Crystal structure of the S.pombe Smc5/6 hinge domain | | Descriptor: | GLYCEROL, SULFATE ION, Structural maintenance of chromosomes protein 5, ... | | Authors: | Alt, A, Pearl, L.H, Oliver, A.W. | | Deposit date: | 2016-11-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Specialized interfaces of Smc5/6 control hinge stability and DNA association.
Nat Commun, 8, 2017
|
|
8OK2
 
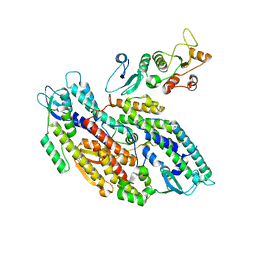 | | Bipartite interaction of TOPBP1 with the GINS complex | | Descriptor: | DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, DNA replication complex GINS protein PSF3, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2023-03-26 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | TopBP1 utilises a bipartite GINS binding mode to support genome replication.
Nat Commun, 15, 2024
|
|
5LOH
 
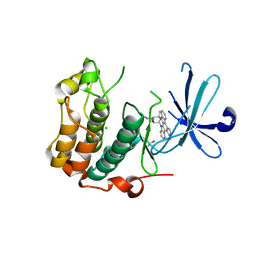 | | Kinase domain of human Greatwall | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, STAUROSPORINE, ... | | Authors: | Rajasekaran, M.B, Pearl, L.H, Oliver, A.W. | | Deposit date: | 2016-08-09 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A first generation inhibitor of human Greatwall kinase, enabled by structural and functional characterisation of a minimal kinase domain construct.
Oncotarget, 7, 2016
|
|
7P0L
 
 | |
7P0J
 
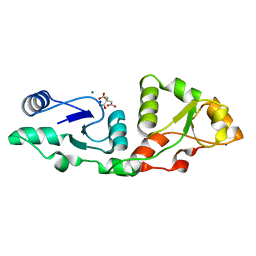 | | Crystal structure of S.pombe Mdb1 BRCT domains | | Descriptor: | CITRIC ACID, DNA damage response protein Mdb1, MAGNESIUM ION, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2021-06-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Phosphorylation-dependent assembly of DNA damage response systems and the central roles of TOPBP1.
DNA Repair (Amst), 108, 2021
|
|
2XM8
 
 | | Co-crystal structure of a small molecule inhibitor bound to the kinase domain of Chk2 | | Descriptor: | 2-{4-[(3S)-PYRROLIDIN-3-YLAMINO]QUINAZOLIN-2-YL}PHENOL, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Caldwell, J.J, Welsh, E.J, Matijssen, C, Anderson, V.E, Antoni, L, Boxall, K, Urban, F, Hayes, A, Raynaud, F.I, Rigoreau, L.J, Raynham, T, Aherne, G.W, Pearl, L.H, Oliver, A.W, Garrett, M.D, Collins, I. | | Deposit date: | 2010-07-26 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-Based Design of Potent and Selective 2-(Quinazolin-2-Yl)Phenol Inhibitors of Checkpoint Kinase 2.
J.Med.Chem., 54, 2011
|
|
2WTC
 
 | | CRYSTAL STRUCTURE OF CHK2 IN COMPLEX WITH AN INHIBITOR | | Descriptor: | 4-[2-AMINO-5-(4-HYDROXY-3-METHOXYPHENYL)PYRIDIN-3-YL]BENZAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Hilton, S, Naud, S, Caldwell, J.J, Boxall, K, Burns, S, Anderson, V.E, Antoni, L, Allen, C.E, Pearl, L.H, Oliver, A.W, Aherne, G.W, Garrett, M.D, Collins, I. | | Deposit date: | 2009-09-15 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification and Characterisation of 2-Aminopyridine Inhibitors of Checkpoint Kinase 2
Bioorg.Med.Chem., 18, 2010
|
|
2XNH
 
 | |
2XNK
 
 | |
2XBJ
 
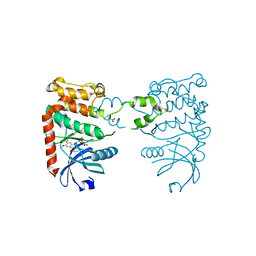 | | Crystal Structure of Chk2 in complex with an inhibitor | | Descriptor: | 4-FLUORO-2-(4-{[(3S,4R)-4-(1-HYDROXY-1-METHYLETHYL)PYRROLIDIN-3-YL]AMINO}-6,7-DIMETHOXYQUINAZOLIN-2-YL)PHENOL, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Anderson, V.E, Walton, M.I, Eve, P.D, Caldwell, J.J, Pearl, L.H, Oliver, A.W, Collins, I, Garrett, M.D. | | Deposit date: | 2010-04-12 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent and Selective 2-(Quinazolin-2-Yl)Phenol Inhibitors of Checkpoint Kinase 2.
J.Med.Chem., 54, 2011
|
|
2XM9
 
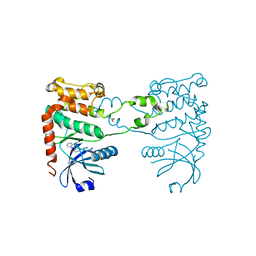 | | Structure of a small molecule inhibitor with the kinase domain of Chk2 | | Descriptor: | 4-(1H-pyrazol-5-yl)-2-{4-[(3S)-pyrrolidin-3-ylamino]quinazolin-2-yl}phenol, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Caldwell, J.J, Welsh, E.J, Matijssen, C, Anderson, V.E, Antoni, L, Boxall, K, Urban, F, Hayes, A, Raynaud, F.I, Rigoreau, L.J, Raynham, T, Aherne, G.W, Pearl, L.H, Oliver, A.W, Garrett, M.D, Collins, I. | | Deposit date: | 2010-07-26 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Potent and Selective 2-(Quinazolin-2-Yl)Phenol Inhibitors of Checkpoint Kinase 2.
J.Med.Chem., 54, 2011
|
|
2WTI
 
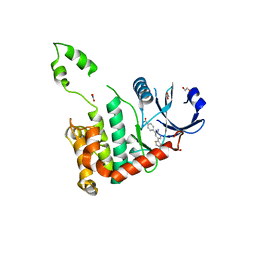 | | CRYSTAL STRUCTURE OF CHK2 IN COMPLEX WITH AN INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-AMINO-5-(2,3-DIHYDROTHIENO[3,4-B][1,4]DIOXIN-5-YL)PYRIDIN-3-YL]BENZAMIDE, CHECKPOINT KINASE 2, ... | | Authors: | Hilton, S, Naud, S, Caldwell, J.J, Boxall, K, Burns, S, Anderson, V.E, Antoni, L, Allen, C.E, Pearl, L.H, Oliver, A.W, Aherne, G.W, Garrett, M.D, Collins, I. | | Deposit date: | 2009-09-16 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification and Characterisation of 2-Aminopyridine Inhibitors of Checkpoint Kinase 2
Bioorg.Med.Chem., 18, 2010
|
|
2WTD
 
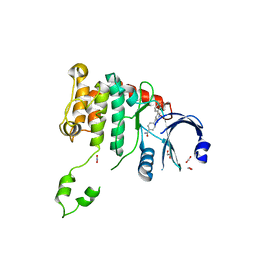 | | Crystal structure of Chk2 in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-AMINO-5-(1,3-BENZODIOXOL-4-YL)PYRIDIN-3-YL]BENZAMIDE, NITRATE ION, ... | | Authors: | Hilton, S, Naud, S, Caldwell, J.J, Boxall, K, Burns, S, Anderson, V.E, Antoni, L, Allen, C.E, Pearl, L.H, Oliver, A.W, Aherne, G.W, Garrett, M.D, Collins, I. | | Deposit date: | 2009-09-15 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification and Characterisation of 2-Aminopyridine Inhibitors of Checkpoint Kinase 2
Bioorg.Med.Chem., 18, 2010
|
|
2WTJ
 
 | | CRYSTAL STRUCTURE OF CHK2 IN COMPLEX WITH AN INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5-(2,3-DIHYDROTHIENO[3,4-B][1,4]DIOXIN-5-YL)-N-[2-(DIMETHYLAMINO)ETHYL]PYRIDINE-3-CARBOXAMIDE, CHECKPOINT KINASE 2, ... | | Authors: | Hilton, S, Naud, S, Caldwell, J.J, Boxall, K, Burns, S, Anderson, V.E, Antoni, L, Allen, C.E, Pearl, L.H, Oliver, A.W, Aherne, G.W, Garrett, M.D, Collins, I. | | Deposit date: | 2009-09-16 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Characterisation of 2-Aminopyridine Inhibitors of Checkpoint Kinase 2
Bioorg.Med.Chem., 18, 2010
|
|
6HM4
 
 | | Crystal structure of Rad4 BRCT1,2 in complex with a Mdb1 phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA damage response protein Mdb1, ... | | Authors: | Day, M, Rappas, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2018-09-12 | | Release date: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (1.770186 Å) | | Cite: | BRCT domains of the DNA damage checkpoint proteins TOPBP1/Rad4 display distinct specificities for phosphopeptide ligands.
Elife, 7, 2018
|
|
6HM3
 
 | | Crystal structure of Rad4 BRCT1,2 in complex with a Sld3 phosphopeptide | | Descriptor: | CALCIUM ION, DNA replication regulator sld3, GLYCEROL, ... | | Authors: | Day, M, Rappas, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2018-09-12 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77263618 Å) | | Cite: | BRCT domains of the DNA damage checkpoint proteins TOPBP1/Rad4 display distinct specificities for phosphopeptide ligands.
Elife, 7, 2018
|
|
4BDE
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 6-METHYLQUINAZOLIN-4-AMINE, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDH
 
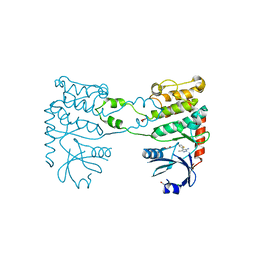 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYL-4-(THIOPHEN-2-YL)-1H-PYRAZOL-5-AMINE, CHLORIDE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDJ
 
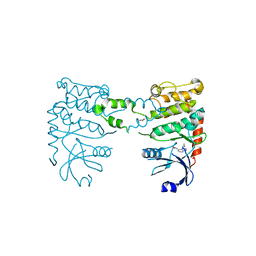 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 3-cyclopropyl-4-(furan-2-yl)-1H-pyrazolo[3,4-b]pyridine, CHECKPOINT KINASE 2, NITRATE ION | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDG
 
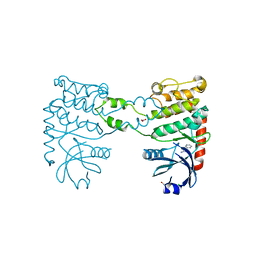 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 3-(PYRIDIN-3-YL)-1H-PYRAZOL-5-AMINE, CHLORIDE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BB7
 
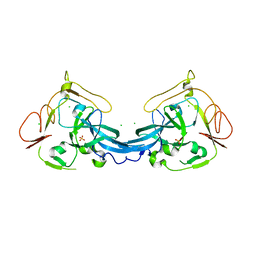 | | Crystal structure of the yeast Rsc2 BAH domain | | Descriptor: | CHLORIDE ION, CHROMATIN STRUCTURE-REMODELING COMPLEX SUBUNIT RSC2, GLYCEROL, ... | | Authors: | Chambers, A.L, Pearl, L.H, Oliver, A.W, Downs, J.A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Bah Domain of Rsc2 is a Histone H3 Binding Domain.
Nucleic Acids Res., 41, 2013
|
|
