2SEM
 
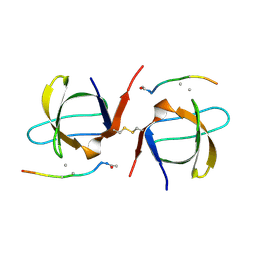 | | SEM5 SH3 DOMAIN COMPLEXED WITH PEPTOID INHIBITOR | | Descriptor: | PROTEIN (SEX MUSCLE ABNORMAL PROTEIN 5), PROTEIN (SH3 PEPTOID INHIBITOR) | | Authors: | Nguyen, J.T, Turck, C.W, Cohen, F.E, Zuckermann, R.N, Lim, W.A. | | Deposit date: | 1998-11-02 | | Release date: | 1999-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploiting the basis of proline recognition by SH3 and WW domains: design of N-substituted inhibitors.
Science, 282, 1998
|
|
4FH8
 
 | |
4H22
 
 | |
1B07
 
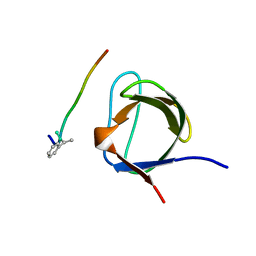 | | CRK SH3 DOMAIN COMPLEXED WITH PEPTOID INHIBITOR | | Descriptor: | PHENYLETHANE, PROTEIN (PROTO-ONCOGENE CRK (CRK)), PROTEIN (SH3 PEPTOID INHIBITOR) | | Authors: | Nguyen, J.T, Turck, C.W, Cohen, F.E, Zuckermann, R.N, Lim, W.A. | | Deposit date: | 1998-11-17 | | Release date: | 1999-01-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Exploiting the basis of proline recognition by SH3 and WW domains: design of N-substituted inhibitors.
Science, 282, 1998
|
|
3SEM
 
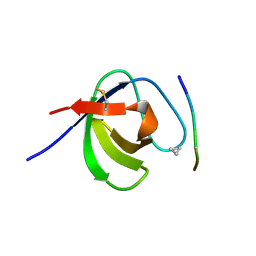 | | SEM5 SH3 DOMAIN COMPLEXED WITH PEPTOID INHIBITOR | | Descriptor: | SEX MUSCLE ABNORMAL PROTEIN 5, SH3 PEPTOID INHIBITOR | | Authors: | Nguyen, J.T, Turck, C.W, Cohen, F.E, Zuckermann, R.N, Lim, W.A. | | Deposit date: | 1998-11-02 | | Release date: | 1999-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploiting the basis of proline recognition by SH3 and WW domains: design of N-substituted inhibitors.
Science, 282, 1998
|
|
4KW6
 
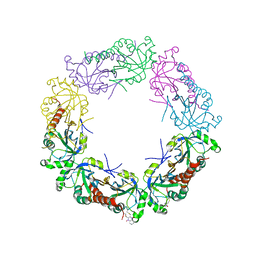 | |
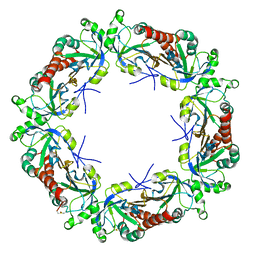
2A4K
 
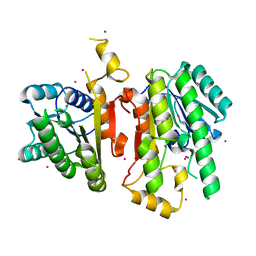 | | 3-Oxoacyl-[acyl carrier protein] reductase from Thermus thermophilus TT0137 | | Descriptor: | 3-oxoacyl-[acyl carrier protein] reductase, UNKNOWN ATOM OR ION | | Authors: | Zhou, W, Ebihara, A, Tempel, W, Yokoyama, S, Chen, L, Kuramitsu, S, Nguyen, J, Chang, S.-H, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-29 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 3-Oxoacyl-[acyl carrier protein] reductase from Thermus thermophilus TT0137
To be Published
|
|
4DT5
 
 | | Crystal Structure of Rhagium inquisitor Antifreeze Protein | | Descriptor: | Antifreeze protein, GLYCEROL, SULFATE ION | | Authors: | Meng, W, Nguyen, J.B, Hakim, A, Thakral, D, Zhu, D.F. | | Deposit date: | 2012-02-20 | | Release date: | 2013-03-13 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of an insect antifreeze protein and its implications for ice binding.
J.Biol.Chem., 288, 2013
|
|
3LIN
 
 | | crystal structure of HTLV protease complexed with the inhibitor, KNI-10562 | | Descriptor: | N-[(2S,3S)-4-{(4R)-4-[(2,2-dimethylpropyl)carbamoyl]-5,5-dimethyl-1,3-thiazolidin-3-yl}-3-hydroxy-4-oxo-1-phenylbutan-2-yl]-N~2~-{(2S)-2-[(methoxycarbonyl)amino]-2-phenylacetyl}-3-methyl-L-valinamide, Protease | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
3LIQ
 
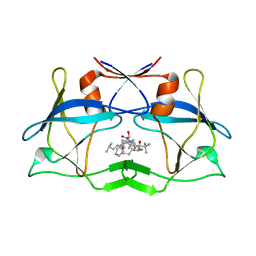 | | Crystal Structure of HTLV protease complexed with the inhibitor, KNI-10673 | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2S)-2-[[(2S)-2-azanyl-2-phenyl-ethanoyl]amino]-3,3-dimethyl-butanoyl]amino]-2-hydroxy-4-phenyl-but anoyl]-5,5-dimethyl-N-(2-methylpropyl)-1,3-thiazolidine-4-carboxamide, Protease | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
3LIY
 
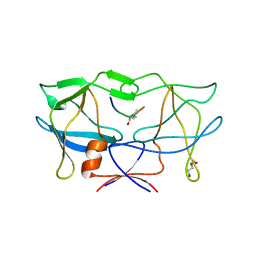 | | crystal structure of HTLV protease complexed with Statine-containing peptide inhibitor | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protease, statine-containing inhibitor | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
3LIX
 
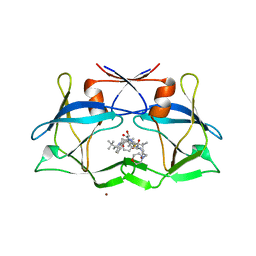 | | crystal structure of htlv protease complexed with the inhibitor KNI-10729 | | Descriptor: | N-{(1S,2S)-1-benzyl-3-[(4R)-5,5-dimethyl-4-{[(1R)-1,2,2-trimethylpropyl]carbamoyl}-1,3-thiazolidin-3-yl]-2-hydroxy-3-oxopropyl}-3-methyl-N~2~-{(2S)-2-[(morpholin-4-ylacetyl)amino]-2-phenylacetyl}-L-valinamide, Protease, ZINC ION | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
3LIV
 
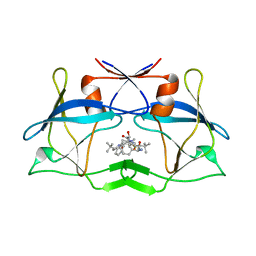 | | crystal structure of HTLV protease complexed with the inhibitor KNI-10683 | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2S)-2-[[(2S)-2-azanyl-2-phenyl-ethanoyl]amino]-3,3-dimethyl-butanoyl]amino]-2-hydroxy-4-phenyl-but anoyl]-N-[(2R)-3,3-dimethylbutan-2-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, Protease | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
3LIT
 
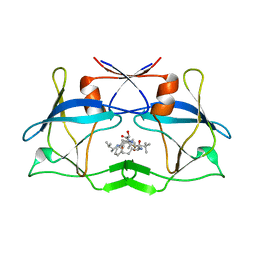 | | The crystal structure of htlv protease complexed with the inhibitor KNI-10681 | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2S)-2-[[(2S)-2-azanyl-2-phenyl-ethanoyl]amino]-3,3-dimethyl-butanoyl]amino]-2-hydroxy-4-phenyl-but anoyl]-5,5-dimethyl-N-[(2R)-3-methylbutan-2-yl]-1,3-thiazolidine-4-carboxamide, Protease | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
4DLQ
 
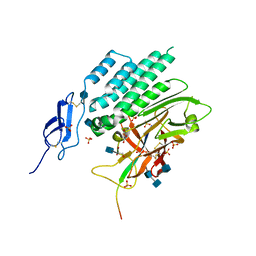 | | Crystal structure of the GAIN and HormR domains of CIRL 1/Latrophilin 1 (CL1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Arac, D, Boucard, A.A, Bolliger, M.F, Nguyen, J, Soltis, M, Sudhof, T.C, Brunger, A.T. | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis.
Embo J., 31, 2012
|
|
6WA0
 
 | |
6W9Y
 
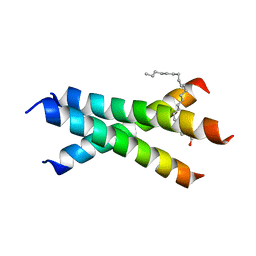 | | De novo designed receptor transmembrane domains enhance CAR-T cytotoxicity and attenuate cytokine release | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, De novo designed receptor transmembrane domain proMP 1.2 | | Authors: | Call, M.J, Call, M.E, Chandler, N.J, Nguyen, J.V, Trenker, R. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | De novo designed receptor transmembrane domains enhance CAR-T cytotoxicity and attenuate cytokine release
To Be Published
|
|
6W9Z
 
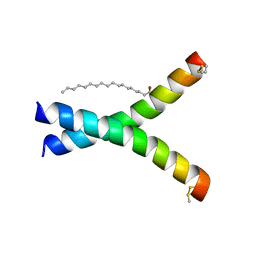 | | De novo designed receptor transmembrane domains enhance CAR-T cytotoxicity and attenuate cytokine release | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, De novo designed receptor transmembrane domain ProMP C2.1 | | Authors: | Call, M.J, Call, M.E, Chandler, N.J, Nguyen, J.V, Trenker, R. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | De novo designed receptor transmembrane domains enhance CAR-T cytotoxicity and attenuate cytokine release
To Be Published
|
|
8VEC
 
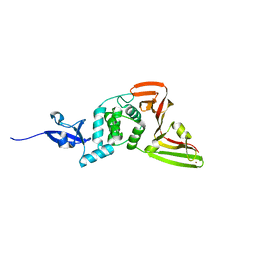 | | Deep Mutational Scanning of SARS-CoV-2 PLpro | | Descriptor: | Papain-like protease nsp3, ZINC ION | | Authors: | Wu, X, Nguyen, J.V, Call, M.E, Call, M.J. | | Deposit date: | 2023-12-18 | | Release date: | 2024-03-20 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational profiling of SARS-CoV-2 papain-like protease reveals requirements for function, structure, and drug escape.
Nat Commun, 15, 2024
|
|
1SHJ
 
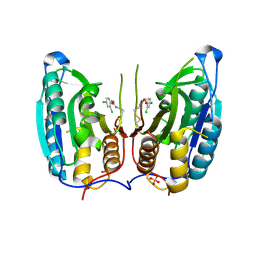 | | Caspase-7 in complex with DICA allosteric inhibitor | | Descriptor: | 2-(2,4-DICHLORO-PHENOXY)-N-(2-MERCAPTO-ETHYL)-ACETAMIDE, Caspase-7, SULFATE ION | | Authors: | Hardy, J.A, Lam, J, Nguyen, J.T, O'Brien, T, Wells, J.A. | | Deposit date: | 2004-02-25 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of an allosteric site in the caspases
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1XHO
 
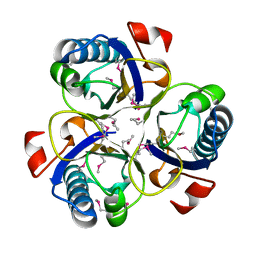 | | Chorismate mutase from Clostridium thermocellum Cth-682 | | Descriptor: | Chorismate mutase, UNKNOWN ATOM OR ION | | Authors: | Xu, H, Chen, L, Lee, D, Habel, J.E, Nguyen, J, Chang, S.-H, Kataeva, I, Chang, J, Zhao, M, Yang, H, Horanyi, P, Florence, Q, Tempel, W, Zhou, W, Lin, D, Zhang, H, Praissman, J, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Ljungdahl, L, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-20 | | Release date: | 2004-11-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Away from the edge II: in-house Se-SAS phasing with chromium radiation.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1SHL
 
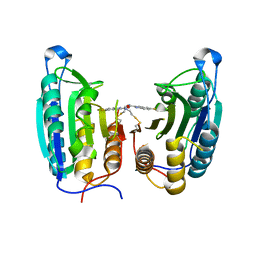 | | CASPASE-7 IN COMPLEX WITH FICA ALLOSTERIC INHIBITOR | | Descriptor: | 5-FLUORO-1H-INDOLE-2-CARBOXYLIC ACID-(2-MERCAPTO-ETHYL)-AMIDE, Caspase-7 | | Authors: | Hardy, J.A, Lam, J, Nguyen, J.T, O'Brien, T, Wells, J.A. | | Deposit date: | 2004-02-25 | | Release date: | 2004-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of an allosteric site in the caspases
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4DLO
 
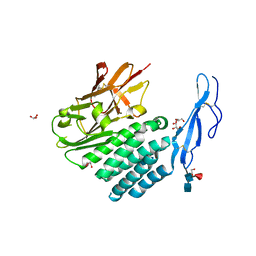 | | Crystal structure of the GAIN and HormR domains of brain angiogenesis inhibitor 3 (BAI3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Brain-specific angiogenesis inhibitor 3, ... | | Authors: | Arac, D, Boucard, A.A, Bolliger, M.F, Nguyen, J, Soltis, M, Sudhof, T.C, Brunger, A.T. | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis.
Embo J., 31, 2012
|
|
4PHU
 
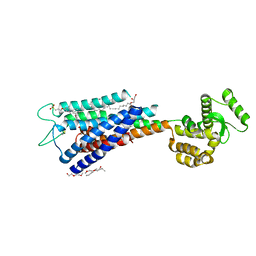 | | Crystal structure of Human GPR40 bound to allosteric agonist TAK-875 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DIMETHYL SULFOXIDE, Free fatty acid receptor 1,Lysozyme, ... | | Authors: | Srivastava, A, Yano, J.K, Hirozane, Y, Kefala, G, Snell, G, Lane, W, Gruswitz, F, Ivetac, A, Aertgeerts, K, Nguyen, J, Jennings, A, Okada, K. | | Deposit date: | 2014-05-07 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.332 Å) | | Cite: | High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875.
Nature, 513, 2014
|
|
1XMA
 
 | | Structure of a transcriptional regulator from Clostridium thermocellum Cth-833 | | Descriptor: | MERCURY (II) ION, Predicted transcriptional regulator, UNKNOWN ATOM OR ION | | Authors: | Yang, H, Chen, L, Lee, D, Habel, J, Nguyen, J, Chang, S.-H, Kataeva, I, Xu, H, Chang, J, Zhao, M, Horanyi, P, Florence, Q, Zhou, W, Tempel, W, Lin, D, Praissman, J, Zhang, H, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Ljungdahl, L, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-10-01 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure of a transcriptional regulator from Clostridium thermocellum Cth-833
To be published
|
|
