4U4Z
 
 | | Crystal structure of Phyllanthoside bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
1OJ5
 
 | | Crystal structure of the Nco-A1 PAS-B domain bound to the STAT6 transactivation domain LXXLL motif | | Descriptor: | IODIDE ION, SIGNAL TRANSDUCER AND ACTIVATOR OF TRANSCRIPTION 6, STEROID RECEPTOR COACTIVATOR 1A | | Authors: | Razeto, A, Ramakrishnan, V, Giller, K, Lakomek, N, Carlomagno, T, Griesinger, C, Lodrini, M, Litterst, C.M, Pftizner, E, Becker, S. | | Deposit date: | 2003-07-02 | | Release date: | 2004-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of the Ncoa-1/Src-1 Pas-B Domain Bound to the Lxxll Motif of the Stat6 Transactivation Domain
J.Mol.Biol., 336, 2004
|
|
2FTE
 
 | | Bacteriophage HK97 Expansion Intermediate IV | | Descriptor: | major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-24 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
7FCI
 
 | | human NTCP in complex with YN69083 Fab | | Descriptor: | Fab Heavy chain, Fab Light chain, Sodium/bile acid cotransporter | | Authors: | Park, J.H, Iwamoto, M, Yun, J.H, Uchikubo-Kamo, T, Son, D, Jin, Z, Yoshida, H, Ohki, M, Ishimoto, N, Mizutani, K, Oshima, M, Muramatsu, M, Wakita, T, Shirouzu, M, Liu, K, Uemura, T, Nomura, N, Iwata, S, Watashi, K, Tame, J.R.H, Nishizawa, T, Lee, W, Park, S.Y. | | Deposit date: | 2021-07-14 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the HBV receptor and bile acid transporter NTCP.
Nature, 606, 2022
|
|
4U3U
 
 | | Crystal structure of Cycloheximide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U4U
 
 | | Crystal structure of Lycorine bound to the yeast 80S ribosome | | Descriptor: | (1S,2S,12bS,12cS)-2,4,5,7,12b,12c-hexahydro-1H-[1,3]dioxolo[4,5-j]pyrrolo[3,2,1-de]phenanthridine-1,2-diol, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
1OIS
 
 | | YEAST DNA TOPOISOMERASE I, N-TERMINAL FRAGMENT | | Descriptor: | DNA TOPOISOMERASE I | | Authors: | Lue, N, Sharma, A, Mondragon, A, Wang, J.C. | | Deposit date: | 1996-09-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 26 kDa yeast DNA topoisomerase I fragment: crystallographic structure and mechanistic implications.
Structure, 3, 1995
|
|
2VOA
 
 | | Structure of an AP Endonuclease from Archaeoglobus fulgidus | | Descriptor: | 5'-D(*CP*GP*GP*CP*TP*AP*CP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*GP*TP*AP*GP*CP*CP*GP)-3', EXODEOXYRIBONUCLEASE III | | Authors: | Kuettner, E.B, Schmiedel, R, Greiner-Stoffele, T, Strater, N. | | Deposit date: | 2008-02-11 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Function of the Abasic Site Specificity Pocket of an Ap Endonuclease from Archaeoglobus Fulgidus.
DNA Repair, 8, 2009
|
|
1OM0
 
 | | crystal structure of xylanase inhibitor protein (XIP-I) from wheat | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Xylanase Inhibitor Protein I | | Authors: | Payan, F, Flatman, R, Porciero, S, Williamson, G, Juge, N, Roussel, A. | | Deposit date: | 2003-02-24 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of xylanase inhibitor protein I (XIP-I), a proteinaceous xylanase inhibitor from wheat (Triticum aestivum, var. Soisson).
Biochem.J., 372, 2003
|
|
1ONK
 
 | | Mistletoe lectin I from viscum album | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AZIDE ION, Beta-galactoside specific lectin I A chain, ... | | Authors: | Gabdoulkhakov, A.G, Savoshkina, Y, Krauspenhaar, R, Stoeva, S, Konareva, N, Kornilov, V, Kornev, A.N, Voelter, W, Nikonov, S.V, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2003-02-28 | | Release date: | 2004-02-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mistletoe lectin I from viscum album
To be Published
|
|
4U6F
 
 | | Crystal structure of T-2 toxin bound to the yeast 80S ribosome | | Descriptor: | 12,13-Epoxytrichothec-9-ene-3,4,8,15-tetrol-4,15-diacetate-8-isovalerate, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-28 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U51
 
 | | Crystal structure of Narciclasine bound to the yeast 80S ribosome | | Descriptor: | (2S,3R,4S,4aR)-2,3,4,7-tetrahydroxy-3,4,4a,5-tetrahydro[1,3]dioxolo[4,5-j]phenanthridin-6(2H)-one, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
1ORO
 
 | | A FLEXIBLE LOOP AT THE DIMER INTERFACE IS A PART OF THE ACTIVE SITE OF THE ADJACENT MONOMER OF ESCHERICHIA COLI OROTATE PHOSPHORIBOSYLTRANSFERASE | | Descriptor: | OROTATE PHOSPHORIBOSYLTRANSFERASE, SULFATE ION | | Authors: | Henriksen, A, Aghajari, N, Jensen, K.F, Gajhede, M. | | Deposit date: | 1995-09-11 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A flexible loop at the dimer interface is a part of the active site of the adjacent monomer of Escherichia coli orotate phosphoribosyltransferase.
Biochemistry, 35, 1996
|
|
1ONU
 
 | | NMDA RECEPTOR ANTAGONIST, CONANTOKIN-G, NMR, 17 STRUCTURES | | Descriptor: | CONANTOKIN-G | | Authors: | Skjaerbaek, N, Nielsen, K.J, Lewis, R.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-08-27 | | Release date: | 1997-09-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of conantokin-G and conantokin-T by CD and NMR spectroscopy.
J.Biol.Chem., 272, 1997
|
|
2J8A
 
 | | X-ray structure of the N-terminus RRM domain of Set1 | | Descriptor: | HISTONE-LYSINE N-METHYLTRANSFERASE, H3 LYSINE-4 SPECIFIC | | Authors: | Tresaugues, L, Dehe, P.M, Guerois, R, Rodriguez-Gil, A, Varlet, I, Salah, P, Pamblanco, M, Luciano, P, Quevillon-Cheruel, S, Sollier, J, Leulliot, N, Couprie, J, Tordera, V, Zinn-Justin, S, Chavez, S, Van Tilbeurgh, H, Geli, V. | | Deposit date: | 2006-10-24 | | Release date: | 2007-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Structure of the N-Terminus Rrm Domain of Set1
To be Published
|
|
4V48
 
 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the initiation-like state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
1P1W
 
 | |
4V73
 
 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in hybrid pre-translocation state (pre5a) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4U56
 
 | | Crystal structure of Blasticidin S bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
1P4L
 
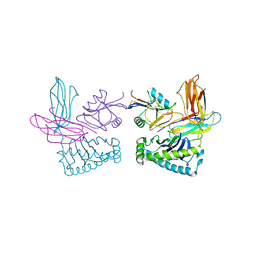 | | Crystal structure of NK receptor Ly49C mutant with its MHC class I ligand H-2Kb | | Descriptor: | Beta-2-microglobulin, LY49-C, MHC CLASS I H-2KB HEAVY CHAIN, ... | | Authors: | Dam, J, Guan, R, Natarajan, K, Dimasi, N, Mariuzza, R.A. | | Deposit date: | 2003-04-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Variable MHC class I engagement by Ly49 natural killer cell receptors demonstrated by the crystal structure of Ly49C bound to H-2K(b).
Nat.Immunol., 4, 2003
|
|
7D2O
 
 | |
4U50
 
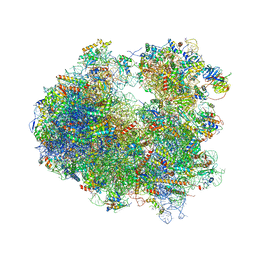 | | Crystal structure of Verrucarin bound to the yeast 80S ribosome | | Descriptor: | (4S,5R,10E,12Z,16R,16aS,17S,18R,19aR,23aR)-4-hydroxy-5,16a,21-trimethyl-4,5,6,7,16,16a,22,23-octahydro-3H,18H,19aH-spiro[16,18-methano[1,6,12]trioxacyclooctadecino[3,4-d]chromene-17,2'-oxirane]-3,9,14-trione, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U3N
 
 | | Crystal structure of CCA trinucleotide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U55
 
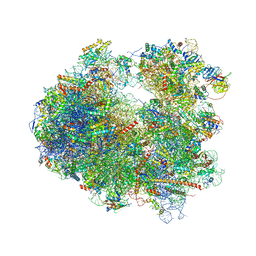 | | Crystal structure of Cryptopleurine bound to the yeast 80S ribosome | | Descriptor: | (14aR)-2,3,6-trimethoxy-11,12,13,14,14a,15-hexahydro-9H-dibenzo[f,h]pyrido[1,2-b]isoquinoline, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
1M6I
 
 | | Crystal Structure of Apoptosis Inducing Factor (AIF) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Programmed cell death protein 8 | | Authors: | Ye, H, Cande, C, Stephanou, N.C, Jiang, S, Gurbuxani, S, Larochette, N, Daugas, E, Garrido, C, Kroemer, G, Wu, H. | | Deposit date: | 2002-07-16 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA binding is required for the apoptogenic action of apoptosis inducing factor.
Nat.Struct.Biol., 9, 2002
|
|
