6G9S
 
 | | Structural basis for the inhibition of E. coli PBP2 | | Descriptor: | (3~{R},6~{S})-6-(aminomethyl)-4-(1,3-oxazol-5-yl)-3-(sulfooxyamino)-3,6-dihydro-2~{H}-pyridine-1-carboxylic acid, Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Ruff, M, Levy, N. | | Deposit date: | 2018-04-11 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
6UYD
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2mc3-v1 redesigned core from genotype 1a bound to broadly neutralizing antibody AR3C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Zhu, J. | | Deposit date: | 2019-11-13 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Proof of concept for rational design of hepatitis C virus E2 core nanoparticle vaccines.
Sci Adv, 6, 2020
|
|
6FOS
 
 | | Cyanidioschyzon merolae photosystem I | | Descriptor: | BETA-CAROTENE, CHLOROPHYLL A, IRON/SULFUR CLUSTER, ... | | Authors: | Nelson, N, Hippler, M, Antoshvili, M, Caspy, I. | | Deposit date: | 2018-02-08 | | Release date: | 2018-04-11 | | Last modified: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure and function of photosystem I in Cyanidioschyzon merolae.
Photosyn. Res., 139, 2019
|
|
6CRX
 
 | | SARS Spike Glycoprotein, Stabilized variant, two S1 CTDs in the upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6OZ7
 
 | | Putative oxidoreductase from Escherichia coli str. K-12 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Osipiuk, J, Skarina, T, Mesa, N, Endres, M, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-15 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Putative oxidoreductase from Escherichia coli str. K-12
to be published
|
|
6FSE
 
 | | Mus musculus acetylcholinesterase in complex with 1-(4-(4-Ethylpiperazin-1-yl)piperidin-1-yl)-2-((4'-methoxy-[1,1'-biphenyl]-4-yl)oxy)ethanone dihydrochloride (15) | | Descriptor: | 1-[4-(4-ethylpiperazin-1-yl)piperidin-1-yl]-2-[4-(4-methoxyphenyl)phenoxy]ethanone, 2-(2-METHOXYETHOXY)ETHANOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ... | | Authors: | Knutsson, S, Engdahl, C, Kumari, R, Kindahl, T, Forsgren, N, Lindgren, C, Kitur, S, Kamau, L, Ekstrom, F, Linusson, A. | | Deposit date: | 2018-02-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Noncovalent Inhibitors of Mosquito Acetylcholinesterase 1 with Resistance-Breaking Potency.
J.Med.Chem., 61, 2018
|
|
6P00
 
 | | RebH Variant 8F, Tryptamine 6-halogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent tryptophan halogenase RebH | | Authors: | Andorfer, M.C, Sukumar, N, Lewis, J.C. | | Deposit date: | 2019-05-16 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Structural and Computational Analysis of Laboratory-Evolved Halogenases Reveals Molecular Details of Site-Selectivity
To Be Published
|
|
5CU5
 
 | | Crystal structure of ERAP2 without catalytic Zn(II) atom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 2, ... | | Authors: | Saridakis, E, Mathioudakis, N, Giastas, P, Mavridis, I.M, Stratikos, E. | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural Basis for Antigenic Peptide Recognition and Processing by Endoplasmic Reticulum (ER) Aminopeptidase 2.
J.Biol.Chem., 290, 2015
|
|
6CUL
 
 | | PvdF of pyoverdin biosynthesis is a structurally unique N10-formyltetrahydrofolate-dependent formyltransferase | | Descriptor: | CITRIC ACID, N-(4-{[(2-amino-4-oxo-1,4-dihydroquinazolin-6-yl)methyl]amino}benzene-1-carbonyl)-D-glutamic acid, Pyoverdine synthetase F | | Authors: | Kenjic, N, Hoag, M.R, Moraski, G.C, Caperelli, C.A, Moran, G.R, Lamb, A.L. | | Deposit date: | 2018-03-26 | | Release date: | 2019-02-06 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | PvdF of pyoverdin biosynthesis is a structurally unique N10-formyltetrahydrofolate-dependent formyltransferase.
Arch. Biochem. Biophys., 664, 2019
|
|
6PPJ
 
 | | Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP | | Descriptor: | ATP-dependent DNA helicase (UvrD/REP), IRON/SULFUR CLUSTER, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | Deposit date: | 2019-07-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6FSN
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with UDP-glucose (conformation 1) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Roversi, P, Le Cornu, J.D, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2018-02-19 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Crystal polymorphism in fragment-based lead discovery of ligands of the catalytic domain of UGGT, the glycoprotein folding quality control checkpoint.
Front Mol Biosci, 2022
|
|
6VA7
 
 | |
6FUJ
 
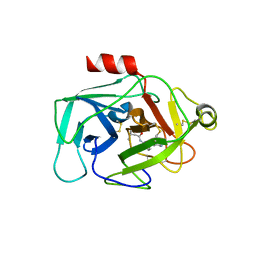 | | Complement factor D in complex with the inhibitor N-(3'-(aminomethyl)-[1,1'-biphenyl]-3-yl)-3-methylbutanamide | | Descriptor: | Complement factor D, ~{N}-[3-[3-(aminomethyl)phenyl]phenyl]-3-methyl-butanamide | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
5D9A
 
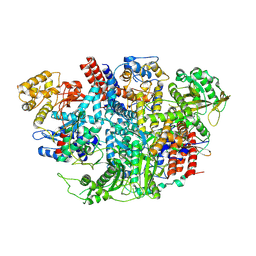 | | Influenza C Virus RNA-dependent RNA Polymerase - Space group P212121 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Hengrung, N, El Omari, K, Serna Martin, I, Vreede, F.T, Cusack, S, Rambo, R.P, Vonrhein, C, Bricogne, G, Stuart, D.I, Grimes, J.M, Fodor, E. | | Deposit date: | 2015-08-18 | | Release date: | 2015-10-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase from influenza C virus.
Nature, 527, 2015
|
|
6P6Y
 
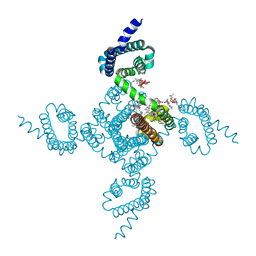 | | Crystal structure of voltage-gated sodium channel NavAb V100C/Q150C disulfide crosslinked mutant in the activated state | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein | | Authors: | Wisedchaisri, G, Tonggu, L, McCord, E, Gamal El-din, T.M, Wang, L, Zheng, N, Catterall, W.A. | | Deposit date: | 2019-06-04 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Resting-State Structure and Gating Mechanism of a Voltage-Gated Sodium Channel.
Cell, 178, 2019
|
|
6G1U
 
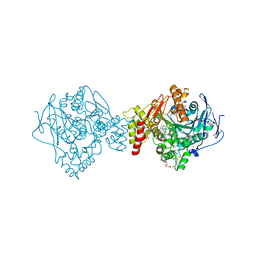 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with 9-Amino-6-chloro-1,2,3,4-tetrahydro-10-methylacridin-10-ium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloranyl-10-methyl-1,2,3,4-tetrahydroacridin-10-ium-9-amine, Acetylcholinesterase, ... | | Authors: | Coquelle, N, Colletier, J.P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Increasing Polarity in Tacrine and Huprine Derivatives: Potent Anticholinesterase Agents for the Treatment of Myasthenia Gravis.
Molecules, 23, 2018
|
|
5D57
 
 | | In meso X-ray crystallography structure of diacylglycerol kinase, DgkA, at 100 K | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Diacylglycerol kinase | | Authors: | Huang, C.-Y, Howe, N, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6CRW
 
 | | SARS Spike Glycoprotein, Stabilized variant, single upwards S1 CTD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6G9F
 
 | | Structural basis for the inhibition of E. coli PBP2 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Ruff, M, Levy, N. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
5D9D
 
 | | Luciferin-regenerating enzyme solved by SAD using synchrotron radiation at room temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
6D2W
 
 | | Crystal structure of Prevotella bryantii endo-beta-mannanase/endo-beta-glucanase PbGH26A-GH5A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aryl-phospho-beta-D-glucosidase BglC, GH1 family, ... | | Authors: | Stogios, P.J, Skarina, T, McGregor, N, Nocek, B, Di Leo, R, Brumer, H, Savchenko, A. | | Deposit date: | 2018-04-14 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Prevotella bryantii endo-beta-mannanase/endo-beta-glucanase PbGH26A-GH5A
To Be Published
|
|
6UYK
 
 | | Dark-operative protochlorophyllide oxidoreductase in the nucleotide-free form. | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase iron-sulfur ATP-binding protein | | Authors: | Bacik, J.P, Imran, S.M.S, Watkins, M.B, Corless, E, Antony, E, Ando, N. | | Deposit date: | 2019-11-13 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The flexible N-terminus of BchL autoinhibits activity through interaction with its [4Fe-4S] cluster and released upon ATP binding.
J.Biol.Chem., 296, 2020
|
|
6UZT
 
 | | Crystal Structure of RPTP alpha | | Descriptor: | Receptor-type tyrosine-protein phosphatase alpha | | Authors: | Santelli, E, Wen, Y, Yang, S, Svensson, M.N.D, Stanford, S.M, Bottini, N. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RPTP alpha phosphatase activity is allosterically regulated by the membrane-distal catalytic domain.
J.Biol.Chem., 295, 2020
|
|
6CMY
 
 | | Solution NMR Structure Determination of Mouse Melanoregulin | | Descriptor: | Melanoregulin | | Authors: | Rout, A.K, Wu, X, Strub, M.P, Starich, M.R, Hammer III, J.A, Tjandra, N. | | Deposit date: | 2018-03-06 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of Melanoregulin Reveals a Role for Cholesterol Recognition in the Protein's Ability to Promote Dynein Function.
Structure, 26, 2018
|
|
6V08
 
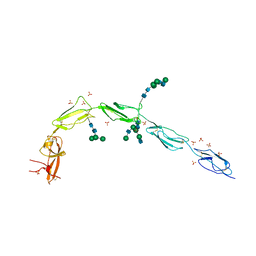 | | Crystal structure of human recombinant Beta-2 glycoprotein I (hrB2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-glycoprotein 1, SULFATE ION, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
