6Z9P
 
 | | Transcription termination intermediate complex 1 | | 分子名称: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | 著者 | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | 登録日 | 2020-06-04 | | 公開日 | 2020-11-04 | | 最終更新日 | 2021-02-03 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6Z8F
 
 | | Human Picobirnavirus D45-CP VLP | | 分子名称: | Capsid protein precursor | | 著者 | Ortega-Esteban, A, Mata, C.P, Rodriguez-Espinosa, M.J, Luque, D, Irigoyen, N, Rodriguez, J.M, de Pablo, P.J, Caston, J.R. | | 登録日 | 2020-06-02 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Cryo-electron Microscopy Structure, Assembly, and Mechanics Show Morphogenesis and Evolution of Human Picobirnavirus.
J.Virol., 94, 2020
|
|
6ZFN
 
 | | Structure of an inactive E404Q variant of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with 1-methyl alpha-1,2-mannobiose | | 分子名称: | Glycoprotein endo-alpha-1,2-mannosidase, SULFATE ION, alpha-D-mannopyranose-(1-2)-methyl alpha-D-mannopyranoside | | 著者 | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | 登録日 | 2020-06-17 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6MUT
 
 | | Cryo-EM structure of ternary Csm-crRNA-target RNA with anti-tag sequence complex in type III-A CRISPR-Cas system | | 分子名称: | RNA (5'-R(*GP*UP*GP*GP*AP*AP*AP*GP*GP*CP*GP*GP*GP*CP*AP*GP*AP*GP*GP*C)-3'), RNA (5'-R(P*CP*CP*UP*CP*UP*GP*CP*CP*CP*GP*CP*CP*UP*UP*UP*CP*CP*AP*C)-3'), Uncharacterized protein Csm1, ... | | 著者 | Jia, N, Wang, C, Eng, E.T, Patel, D.J. | | 登録日 | 2018-10-23 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
6Z9H
 
 | | Escherichia coli D-2-deoxyribose-5-phosphate aldolase - C47V/G204A/S239D mutant | | 分子名称: | 1,2-ETHANEDIOL, Deoxyribose-phosphate aldolase, FORMIC ACID, ... | | 著者 | Paakkonen, J, Hakulinen, N, Rouvinen, J. | | 登録日 | 2020-06-04 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Substrate specificity of 2-deoxy-D-ribose 5-phosphate aldolase (DERA) assessed by different protein engineering and machine learning methods.
Appl.Microbiol.Biotechnol., 104, 2020
|
|
1F9B
 
 | | MELANIN PROTEIN INTERACTION: X-RAY STRUCTURE OF THE COMPLEX OF MARE LACTOFERRIN WITH MELANIN MONOMERS | | 分子名称: | 3H-INDOLE-5,6-DIOL, BICARBONATE ION, FE (III) ION, ... | | 著者 | Kumar, S, Singh, T.P, Sharma, A.K, Singh, N, Raman, G. | | 登録日 | 2000-07-10 | | 公開日 | 2001-02-10 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Lactoferrin-melanin interaction and its possible implications in melanin polymerization: crystal structure of the complex formed between mare lactoferrin and melanin monomers at 2.7-A resolution.
Proteins, 45, 2001
|
|
4ETC
 
 | | Lysozyme, room temperature, 24 kGy dose | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | 登録日 | 2012-04-24 | | 公開日 | 2012-06-13 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.906 Å) | | 主引用文献 | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
6YSR
 
 | | Structure of the P+9 stalled ribosome complex | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | 登録日 | 2020-04-23 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
2VRA
 
 | | Drosophila Robo IG1-2 (monoclinic form) | | 分子名称: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ROUNDABOUT 1, SULFATE ION | | 著者 | Fukuhara, N, Howitt, J.A, Hussain, S, Hohenester, E. | | 登録日 | 2008-03-28 | | 公開日 | 2008-04-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural and Functional Analysis of Slit and Heparin Binding to Immunoglobulin-Like Domains 1 and 2 of Drosophila Robo
J.Biol.Chem., 283, 2008
|
|
6YXQ
 
 | | Crystal structure of a DNA repair complex ASCC3-ASCC2 | | 分子名称: | Activating signal cointegrator 1 complex subunit 2, Activating signal cointegrator 1 complex subunit 3 | | 著者 | Jia, J, Absmeier, E, Holton, N, Bohnsack, K.E, Pietrzyk-Brzezinska, A.J, Bohnsack, M.T, Wahl, M.C. | | 登録日 | 2020-05-03 | | 公開日 | 2020-09-30 | | 最終更新日 | 2020-11-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The interaction of DNA repair factors ASCC2 and ASCC3 is affected by somatic cancer mutations.
Nat Commun, 11, 2020
|
|
6MU5
 
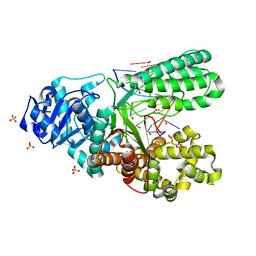 | | Bst DNA polymerase I TNA/DNA binary complex | | 分子名称: | DNA (5'-D(P*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA polymerase I, SULFATE ION, ... | | 著者 | Jackson, L.N, Chim, N, Chaput, J.C. | | 登録日 | 2018-10-22 | | 公開日 | 2019-06-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.912 Å) | | 主引用文献 | Crystal structures of a natural DNA polymerase that functions as an XNA reverse transcriptase.
Nucleic Acids Res., 47, 2019
|
|
1F7Z
 
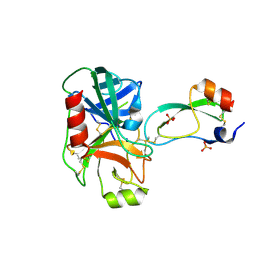 | | RAT TRYPSINOGEN K15A COMPLEXED WITH BOVINE PANCREATIC TRYPSIN INHIBITOR | | 分子名称: | CALCIUM ION, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION, ... | | 著者 | Pasternak, A, White, A, Jeffery, C.J, Medina, N, Cahoon, M, Ringe, D, Hedstrom, L. | | 登録日 | 2000-06-28 | | 公開日 | 2001-07-04 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The energetic cost of induced fit catalysis: Crystal structures of trypsinogen mutants with enhanced activity and inhibitor affinity.
Protein Sci., 10, 2001
|
|
1F7Y
 
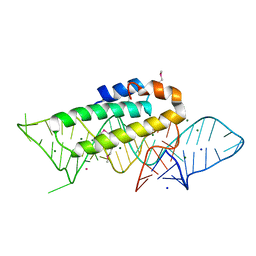 | | THE CRYSTAL STRUCTURE OF TWO UUCG LOOPS HIGHLIGHTS THE ROLE PLAYED BY 2'-HYDROXYL GROUPS IN ITS UNUSUAL STABILITY | | 分子名称: | 16S RIBOSOMAL RNA FRAGMENT, 30S RIBOSOMAL PROTEIN S15, MAGNESIUM ION, ... | | 著者 | Ennifar, E, Nikouline, A, Serganov, A, Tishchenko, S, Nevskaya, N, Garber, M, Ehresmann, B, Ehresmann, C, Nikonov, S, Dumas, P. | | 登録日 | 2000-06-28 | | 公開日 | 2000-11-22 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The crystal structure of UUCG tetraloop.
J.Mol.Biol., 304, 2000
|
|
6MV6
 
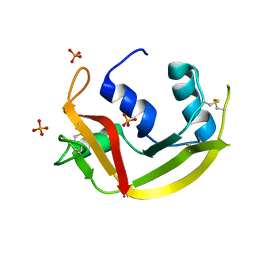 | | Crystal structure of RNAse 6 | | 分子名称: | PHOSPHATE ION, Ribonuclease K6 | | 著者 | Couture, J.-F, Doucet, N. | | 登録日 | 2018-10-24 | | 公開日 | 2019-11-13 | | 最終更新日 | 2020-05-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Insights into Structural and Dynamical Changes Experienced by Human RNase 6 upon Ligand Binding.
Biochemistry, 59, 2020
|
|
6Z8D
 
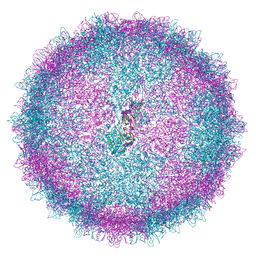 | | Human Picobirnavirus CP VLP | | 分子名称: | Capsid protein precursor | | 著者 | Ortega-Esteban, A, Mata, C.P, Rodriguez-Espinosa, M.J, Luque, D, Irigoyen, N, Rodriguez, J.M, de Pablo, P.J, Caston, J.R. | | 登録日 | 2020-06-02 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.63 Å) | | 主引用文献 | Cryo-electron Microscopy Structure, Assembly, and Mechanics Show Morphogenesis and Evolution of Human Picobirnavirus.
J.Virol., 94, 2020
|
|
6Z9F
 
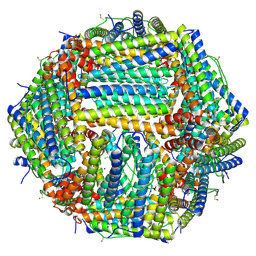 | | 1.56 A structure of human apoferritin obtained from data subset of Titan Mono-BCOR microscope | | 分子名称: | Ferritin heavy chain, SODIUM ION | | 著者 | Yip, K.M, Fischer, N, Paknia, E, Chari, A, Stark, H. | | 登録日 | 2020-06-03 | | 公開日 | 2020-06-24 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (1.56 Å) | | 主引用文献 | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
6ZCA
 
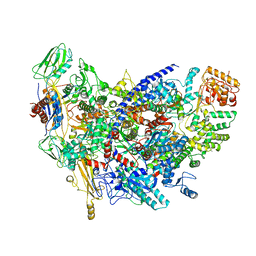 | | Structure of the B. subtilis RNA POLYMERASE in complex with HelD (monomer) | | 分子名称: | DNA helicase, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Pei, H.-P, Hilal, T, Huang, Y.-H, Said, N, Loll, B, Wahl, M.C. | | 登録日 | 2020-06-10 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | The delta subunit and NTPase HelD institute a two-pronged mechanism for RNA polymerase recycling.
Nat Commun, 11, 2020
|
|
6MLQ
 
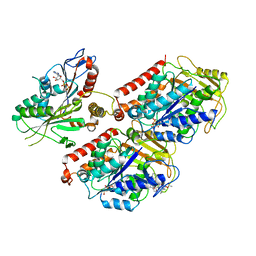 | | Cryo-EM structure of microtubule-bound Kif7 in the ADP state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Mani, N, Jiang, S, Wilson-Kubalek, E.M, Ku, P, Milligan, R.A, Subramanian, R. | | 登録日 | 2018-09-27 | | 公開日 | 2019-05-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Interplay between the Kinesin and Tubulin Mechanochemical Cycles Underlies Microtubule Tip Tracking by the Non-motile Ciliary Kinesin Kif7.
Dev.Cell, 49, 2019
|
|
2VDC
 
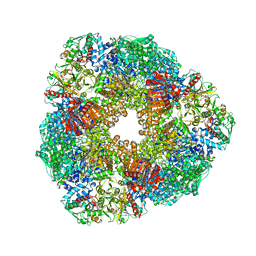 | | THE 9.5 A RESOLUTION STRUCTURE OF GLUTAMATE SYNTHASE FROM CRYO-ELECTRON MICROSCOPY AND ITS OLIGOMERIZATION BEHAVIOR IN SOLUTION: FUNCTIONAL IMPLICATIONS. | | 分子名称: | 2-OXOGLUTARIC ACID, FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Cottevieille, M, Larquet, E, Jonic, S, Petoukhov, M.V, Caprini, G, Paravisi, S, Svergun, D.I, Vanoni, M.A, Boisset, N. | | 登録日 | 2007-10-04 | | 公開日 | 2008-01-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (9.5 Å) | | 主引用文献 | The Subnanometer Resolution Structure of the Glutamate Synthase 1.2-Mda Hexamer by Cryoelectron Microscopy and its Oligomerization Behavior in Solution: Functional Implications.
J.Biol.Chem., 283, 2008
|
|
1FBY
 
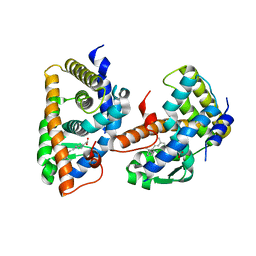 | | CRYSTAL STRUCTURE OF THE HUMAN RXR ALPHA LIGAND BINDING DOMAIN BOUND TO 9-CIS RETINOIC ACID | | 分子名称: | (9cis)-retinoic acid, RETINOIC ACID RECEPTOR RXR-ALPHA | | 著者 | Egea, P.F, Mitschler, A, Rochel, N, Ruff, M, Chambon, P, Moras, D. | | 登録日 | 2000-07-17 | | 公開日 | 2000-07-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structure of the human RXRalpha ligand-binding domain bound to its natural ligand: 9-cis retinoic acid.
EMBO J., 19, 2000
|
|
6ZLM
 
 | | Dihydrolipoyllysine-residue acetyltransferase component of fungal pyruvate dehydrogenase complex with protein X bound | | 分子名称: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial, Pyruvate dehydrogenase X component | | 著者 | Forsberg, B.O, Aibara, S, Howard, R.J, Mortezaei, N, Lindahl, E. | | 登録日 | 2020-06-30 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Arrangement and symmetry of the fungal E3BP-containing core of the pyruvate dehydrogenase complex.
Nat Commun, 11, 2020
|
|
6YTZ
 
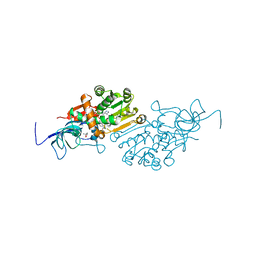 | | Crystal structure of Malus domestica Double Bond Reductase (MdDBR) in complex with NADPH | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Double Bond Reductase, ... | | 著者 | Caliandro, R, Polsinelli, I, Demitri, N, Benini, S. | | 登録日 | 2020-04-25 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The structural and functional characterization of Malus domestica double bond reductase MdDBR provides insights towards the identification of its substrates.
Int.J.Biol.Macromol., 171, 2021
|
|
1FI5
 
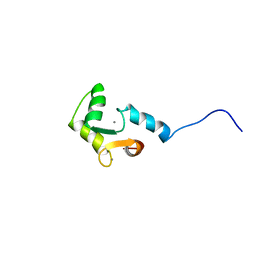 | | NMR STRUCTURE OF THE C TERMINAL DOMAIN OF CARDIAC TROPONIN C BOUND TO THE N TERMINAL DOMAIN OF CARDIAC TROPONIN I. | | 分子名称: | CALCIUM ION, PROTEIN (TROPONIN C) | | 著者 | Gasmi-Seabrook, G.M, Howarth, J.W, Finley, N, Abusamhadneh, E, Gaponenko, V, Brito, R.M, Solaro, R.J, Rosevear, P.R. | | 登録日 | 2000-08-03 | | 公開日 | 2000-08-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures of the C-terminal domain of cardiac troponin C free and bound to the N-terminal domain of cardiac troponin I.
Biochemistry, 38, 1999
|
|
1FBV
 
 | | STRUCTURE OF A CBL-UBCH7 COMPLEX: RING DOMAIN FUNCTION IN UBIQUITIN-PROTEIN LIGASES | | 分子名称: | SIGNAL TRANSDUCTION PROTEIN CBL, SULFATE ION, UBIQUITIN-CONJUGATING ENZYME E12-18 KDA UBCH7, ... | | 著者 | Zheng, N, Wang, P, Jeffrey, P.D, Pavletich, N.P. | | 登録日 | 2000-07-17 | | 公開日 | 2000-08-30 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases.
Cell(Cambridge,Mass.), 102, 2000
|
|
6XJ3
 
 | | Crystal structure of Class D beta-lactamase from Klebsiella quasipneumoniae in complex with avibactam | | 分子名称: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-22 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Class D beta-lactamase from Klebsiella quasipneumoniae
To Be Published
|
|
