5KPM
 
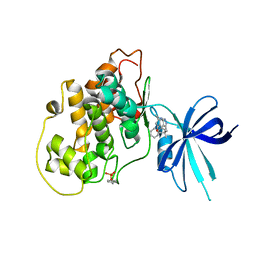 | | Glycogen Synthase Kinase 3 beta Complexed with BRD3731 | | Descriptor: | (4~{S})-3-(2,2-dimethylpropyl)-4,7,7-trimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5KPL
 
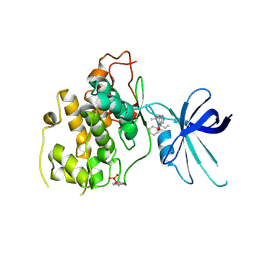 | | Glycogen Synthase Kinase 3 beta Complexed with BRD0705 | | Descriptor: | (4~{S})-4-ethyl-7,7-dimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5KPK
 
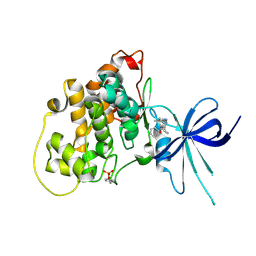 | | Glycogen Synthase Kinase 3 beta Complexed with BRD0209 | | Descriptor: | (4~{S})-3-cyclopropyl-4,7,7-trimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5VZ3
 
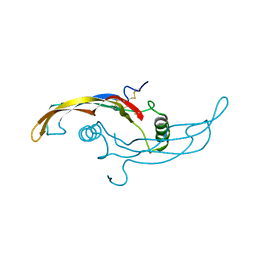 | |
5VZ4
 
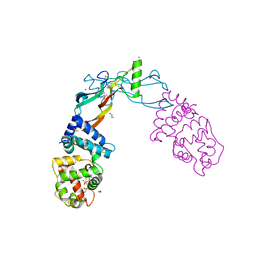 | | Receptor-growth factor crystal structure at 2.20 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, GDNF family receptor alpha-like, ... | | Authors: | Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2017-05-26 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Non-homeostatic body weight regulation through a brainstem-restricted receptor for GDF15.
Nature, 550, 2017
|
|
4YH4
 
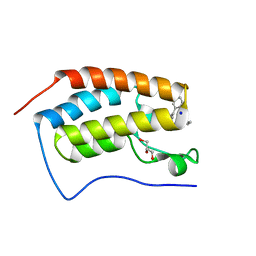 | | Crystal structure of human BRD4(1) in complex with 4-[(5-phenylpyridin-3-yl)carbonyl]-3,4-dihydroquinoxalin-2(1H)-one (compound 19d) | | Descriptor: | 4-[(5-phenylpyridin-3-yl)carbonyl]-3,4-dihydroquinoxalin-2(1H)-one, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of a new chemical series of BRD4(1) inhibitors using protein-ligand docking and structure-guided design.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6CZV
 
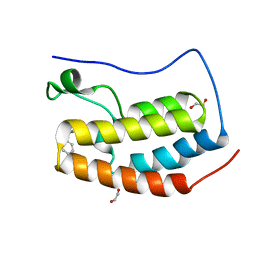 | | BRD4(BD1) complexed with 2759 | | Descriptor: | 1,2-ETHANEDIOL, 1-benzyl-5-(3,5-dimethyl-1,2-oxazol-4-yl)pyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Design and Characterization of Novel Covalent Bromodomain and Extra-Terminal Domain (BET) Inhibitors Targeting a Methionine.
J. Med. Chem., 61, 2018
|
|
6CZU
 
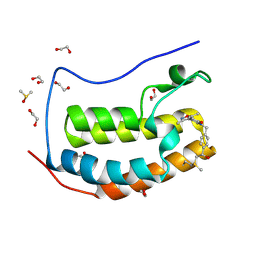 | | BRD4(BD1) complexed with 3219 | | Descriptor: | 1,2-ETHANEDIOL, 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-({4-[(1R)-1-hydroxyethyl]phenyl}methyl)pyridin-2(1H)-one, Bromodomain-containing protein 4, ... | | Authors: | Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-26 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Design and Characterization of Novel Covalent Bromodomain and Extra-Terminal Domain (BET) Inhibitors Targeting a Methionine.
J. Med. Chem., 61, 2018
|
|
4L19
 
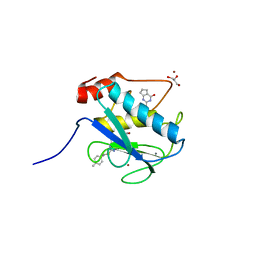 | | Matrix metalloproteinase-13 complexed with selective inhibitor compound Q1 | | Descriptor: | 2-[(4-methylbenzyl)sulfanyl]-3,5,6,7-tetrahydro-4H-cyclopenta[d]pyrimidin-4-one, CALCIUM ION, Collagenase 3, ... | | Authors: | Minond, D, Spicer, T.P, Jiang, J, Taylor, A.B, Hart, P.J, Roush, W.R, Fields, G.B, Hodder, P.S. | | Deposit date: | 2013-06-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Characterization of selective exosite-binding inhibitors of matrix metalloproteinase 13 that prevent articular cartilage degradation in vitro.
J.Med.Chem., 57, 2014
|
|
1QX4
 
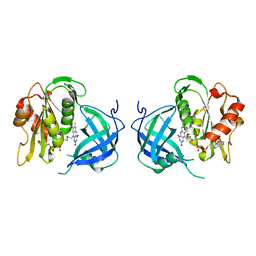 | | Structrue of S127P mutant of cytochrome b5 reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase | | Authors: | Bewley, M.C, Davis, C.A, Marohnic, C.C, Taormina, D, Barber, M.J. | | Deposit date: | 2003-09-04 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the S127P mutant of cytochrome b5 reductase that causes methemoglobinemia shows the AMP moiety of the flavin occupying the substrate binding site
Biochemistry, 42, 2003
|
|
6E4A
 
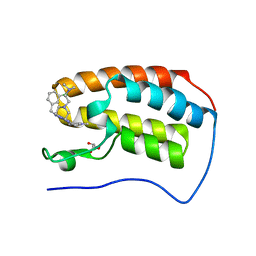 | | Crystal structure of human BRD4(1) in complex with CN750 | | Descriptor: | 5-(4-{[(3-chlorophenyl)methyl]amino}-2-{4-[2-(dimethylamino)ethyl]piperazin-1-yl}quinazolin-6-yl)-1-methylpyridin-2(1H)-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Fontano, E, White, A, Lakshminarasimhan, D, Suto, R.K. | | Deposit date: | 2018-07-17 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery and lead identification of quinazoline-based BRD4 inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
7X4L
 
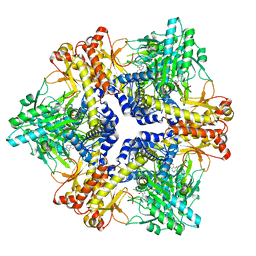 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase mutant Y303F-PLP complex | | Descriptor: | Glutamate decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Liu, S, Guoming, D, Yulu, W, Boting, W, Xin, F. | | Deposit date: | 2022-03-02 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
6B85
 
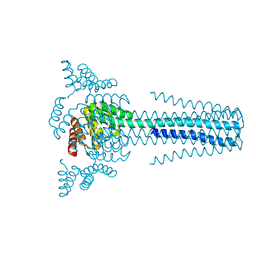 | | Crystal structure of transmembrane protein TMHC4_R | | Descriptor: | TMHC4_R | | Authors: | Lu, P, DiMaio, F, Min, D, Bowie, J, Wei, K.Y, Baker, D. | | Deposit date: | 2017-10-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.889 Å) | | Cite: | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
6B87
 
 | | Crystal structure of transmembrane protein TMHC2_E | | Descriptor: | TMHC2_E | | Authors: | Lu, P, DiMaio, F, Min, D, Wei, K.Y, Bowie, J, Baker, D. | | Deposit date: | 2017-10-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.947 Å) | | Cite: | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
7V5C
 
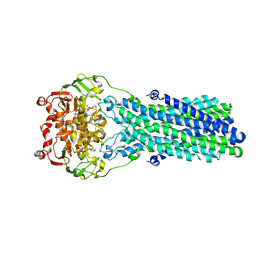 | | Cryo-EM structure of the mouse ABCB9 (ADP.BeF3-bound) | | Descriptor: | ABC-type oligopeptide transporter ABCB9, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Park, J.G, Kim, S, Jang, E, Choi, S.H, Han, H, Kim, J.W, Ju, S, Min, D.S, Jin, M.S. | | Deposit date: | 2021-08-17 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The lysosomal transporter TAPL has a dual role as peptide translocator and phosphatidylserine floppase.
Nat Commun, 13, 2022
|
|
7V5D
 
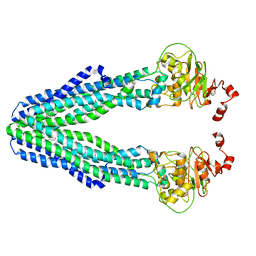 | | Cryo-EM structure of the mouse ABCB9 (PG-bound) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, ABC-type oligopeptide transporter ABCB9 | | Authors: | Park, J.G, Kim, S, Jang, E, Choi, S.H, Han, H, Ju, S, Kim, J.W, Min, D.S, Jin, M.S. | | Deposit date: | 2021-08-17 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The lysosomal transporter TAPL has a dual role as peptide translocator and phosphatidylserine floppase.
Nat Commun, 13, 2022
|
|
7VFI
 
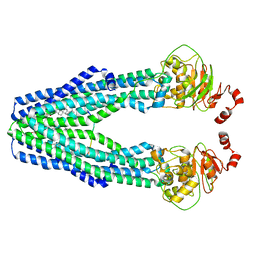 | | Cryo-EM structure of the mouse TAPL (9mer-peptide bound) | | Descriptor: | ABC-type oligopeptide transporter ABCB9, ARG-ARG-TYR-GLN-LYS-SER-THR-GLU-LEU, CHOLESTEROL HEMISUCCINATE | | Authors: | Park, J.G, Kim, S, Jang, E, Choi, S.H, Han, H, Kim, J.W, Ju, S, Min, D.S, Jin, M.S. | | Deposit date: | 2021-09-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | The lysosomal transporter TAPL has a dual role as peptide translocator and phosphatidylserine floppase.
Nat Commun, 13, 2022
|
|
7OZS
 
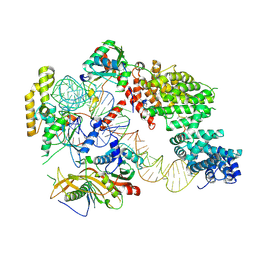 | | Structure of the hexameric 5S RNP from C. thermophilum | | Descriptor: | 5S rRNA, 60S ribosomal protein l5-like protein, Putative ribosomal protein, ... | | Authors: | Castillo, N, Thoms, M, Flemming, D, Hammaren, H.M, Buschauer, R, Ameismeier, M, Bassler, J, Beck, M, Beckmann, R, Hurt, E. | | Deposit date: | 2021-06-28 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of nascent 5S RNPs at the crossroad between ribosome assembly and MDM2-p53 pathways.
Nat.Struct.Mol.Biol., 2023
|
|
6PLG
 
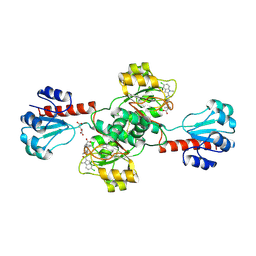 | | Crystal structure of human PHGDH complexed with Compound 15 | | Descriptor: | (2S)-(4-{3-[(4,5-dichloro-1-methyl-1H-indole-2-carbonyl)amino]oxetan-3-yl}phenyl)(pyridin-3-yl)acetic acid, D-3-phosphoglycerate dehydrogenase, D-MALATE | | Authors: | Olland, A, Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2019-06-30 | | Release date: | 2019-07-24 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Inhibition of 3-phosphoglycerate dehydrogenase (PHGDH) by indole amides abrogates de novo serine synthesis in cancer cells.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
5JPQ
 
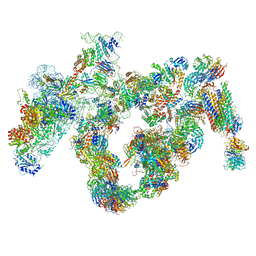 | | Cryo-EM structure of the 90S pre-ribosome | | Descriptor: | 18S ribosomal RNA, Bms1, Emg1, ... | | Authors: | Turk, M, Cheng, J, Berninghausen, O, Kornprobst, M, Flemming, D, Kos-Braun, I.C, Kos, M, Thoms, M, Hurt, E, Beckmann, R. | | Deposit date: | 2016-05-04 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Architecture of the 90S Pre-ribosome: A Structural View on the Birth of the Eukaryotic Ribosome.
Cell, 166, 2016
|
|
6PLF
 
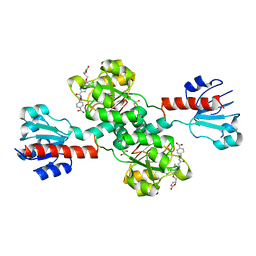 | | Crystal structure of human PHGDH complexed with Compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 4-{(1S)-1-[(5-chloro-6-{[(5S)-2-oxo-1,3-oxazolidin-5-yl]methoxy}-1H-indole-2-carbonyl)amino]-2-hydroxyethyl}benzoic acid, D-3-phosphoglycerate dehydrogenase | | Authors: | Olland, A, Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2019-06-30 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of 3-phosphoglycerate dehydrogenase (PHGDH) by indole amides abrogates de novo serine synthesis in cancer cells.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
4UIR
 
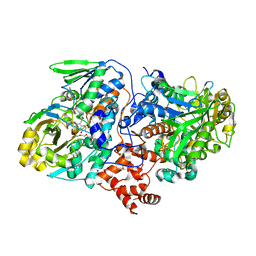 | | Structure of oleate hydratase from Elizabethkingia meningoseptica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HEXAETHYLENE GLYCOL, OLEATE HYDRATASE, ... | | Authors: | Pavkov-Keller, T, Hromic, A, Engleder, M, Emmerstorfer, A, Steinkellner, G, Schrempf, S, Wriessnegger, T, Leitner, E, Strohmeier, G.A, Kaluzna, I, Mink, D, Schuermann, M, Wallner, S, Macheroux, P, Pichler, H, Gruber, K. | | Deposit date: | 2015-04-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Based Mechanism of Oleate Hydratase from Elizabethkingia Meningoseptica.
Chembiochem, 16, 2015
|
|
6WMW
 
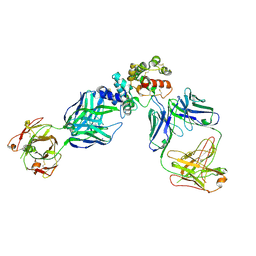 | | GFRAL receptor bound with two antibody Fabs (3P10, 25M22) | | Descriptor: | FAB25M22 heavy chain fragment, FAB25M22 light chain, FAB3P10 heavy chain fragment, ... | | Authors: | White, A, Lakshminarasimhan, D, Olland, A, Suto, R.K. | | Deposit date: | 2020-04-21 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Antibody-mediated inhibition of GDF15-GFRAL activity reverses cancer cachexia in mice.
Nat Med, 26, 2020
|
|
5JCS
 
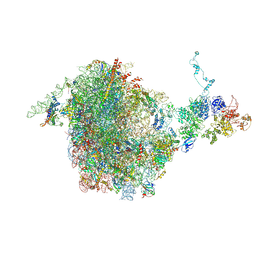 | | CRYO-EM STRUCTURE OF THE RIX1-REA1 PRE-60S PARTICLE | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Barrio-Garcia, C, Thoms, M, Flemming, D, Kater, L, Berninghausen, O, Bassler, J, Beckmann, R, Hurt, E. | | Deposit date: | 2016-04-15 | | Release date: | 2016-11-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Architecture of the Rix1-Rea1 checkpoint machinery during pre-60S-ribosome remodeling
Nat.Struct.Mol.Biol., 23, 2016
|
|
8BGU
 
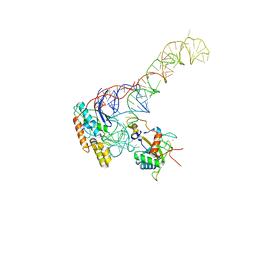 | | human MDM2-5S RNP | | Descriptor: | 5S rRNA, 60S ribosomal protein L11, 60S ribosomal protein L5, ... | | Authors: | Castillo, N, Thoms, M, Flemming, D, Hammaren, H.M, Buschauer, R, Ameismeier, M, Bassler, J, Beck, M, Beckmann, R, Hurt, E. | | Deposit date: | 2022-10-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of nascent 5S RNPs at the crossroad between ribosome assembly and MDM2-p53 pathways.
Nat.Struct.Mol.Biol., 30, 2023
|
|
