3FD7
 
 | | Crystal structure of Onconase C87A/C104A-ONC | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein P-30, ... | | Authors: | Neumann, P, Schulenburg, C, Arnold, U, Ulbrich-Hofmann, R, Stubbs, M.T. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | Impact of the C-terminal disulfide bond on the folding and stability of onconase.
Chembiochem, 11, 2010
|
|
2WL8
 
 | | X-ray crystal structure of Pex19p | | Descriptor: | PEROXISOMAL BIOGENESIS FACTOR 19 | | Authors: | Schueller, N, Holton, S.J, Stanley, W.A, Song, Y.H, Konarev, P, Roessle, M, Erdmann, R, Schliebs, W, Wilmanns, M. | | Deposit date: | 2009-06-22 | | Release date: | 2010-06-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Peroxisomal Receptor Pex19P Forms a Helical Mpts Recognition Domain.
Embo J., 29, 2010
|
|
1XK5
 
 | | Crystal structure of the m3G-cap-binding domain of snurportin1 in complex with a m3GpppG-cap dinucleotide | | Descriptor: | 2,2,7-TRIMETHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, snurportin-1 | | Authors: | Strasser, A, Dickmanns, A, Luehrmann, R, Ficner, R. | | Deposit date: | 2004-09-27 | | Release date: | 2005-06-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for m(3)G-cap-mediated nuclear import of spliceosomal UsnRNPs by snurportin1
Embo J., 24, 2005
|
|
5A6U
 
 | | Native mammalian ribosome-bound Sec61 protein-conducting channel in the 'non-inserting' state | | Descriptor: | SEC61A, SEC61B, SEC61G | | Authors: | Pfeffer, S, Burbaum, L, Unverdorben, P, Pech, M, Chen, Y, Zimmermann, R, Beckmann, R, Foerster, F. | | Deposit date: | 2015-07-01 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of the Native Sec61 Protein-Conducting Channel.
Nat.Commun., 6, 2015
|
|
4V4B
 
 | | Structure of the ribosomal 80S-eEF2-sordarin complex from yeast obtained by docking atomic models for RNA and protein components into a 11.7 A cryo-EM map. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S11, ... | | Authors: | Spahn, C.M, Gomez-Lorenzo, M.G, Grassucci, R.A, Jorgensen, R, Andersen, G.R, Beckmann, R, Penczek, P.A, Ballesta, J.P.G, Frank, J. | | Deposit date: | 2004-01-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation.
Embo J., 23, 2004
|
|
3OC3
 
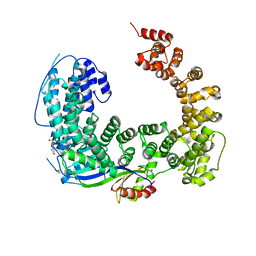 | | Crystal structure of the Mot1 N-terminal domain in complex with TBP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HELICASE MOT1, TRANSCRIPTION INITIATION FACTOR TFIID (TFIID-1) | | Authors: | Wollmann, P, Cui, S, Viswanathan, R, Berninghausen, O, Wells, M.N, Moldt, M, Witte, G, Butryn, A, Wendler, P, Beckmann, R, Auble, D.T, Hopfner, K.-P. | | Deposit date: | 2010-08-09 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP.
Nature, 475, 2011
|
|
4WQ2
 
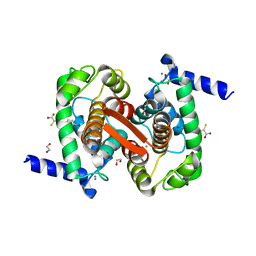 | | Human calpain PEF(S) with (Z)-3-(6-bromondol-3-yl)-2-mercaptoacrylic acid bound | | Descriptor: | (2Z)-3-(6-bromo-1H-indol-3-yl)-2-sulfanylprop-2-enoic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Adams, S.E, Rizkallah, P.J, Miller, D.J, Hallett, M.B, Allemann, R.K. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Conformationally restricted calpain inhibitors.
Chem Sci, 6, 2015
|
|
4WQ3
 
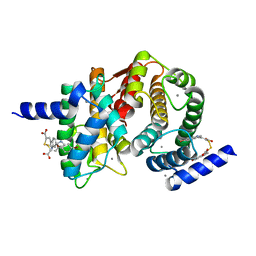 | | Human calpain PEF(S) with (2Z,2Z')-2,2'-disulfanediylbis(3-(6-bromoindol-3-yl)acrylic acid) bound | | Descriptor: | (2Z)-3-(6-bromo-1H-indol-3-yl)-2-sulfanylprop-2-enoic acid, CALCIUM ION, Calpain small subunit 1 | | Authors: | Adams, S.E, Robinson, E.J, Rizkallah, P.J, Miller, D.J, Hallett, M.B, Allemann, R.K. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Conformationally restricted calpain inhibitors.
Chem Sci, 6, 2015
|
|
2FWG
 
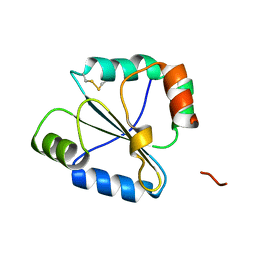 | | high resolution crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (photoreduced form) | | Descriptor: | Thiol:disulfide interchange protein dsbD | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
2FWE
 
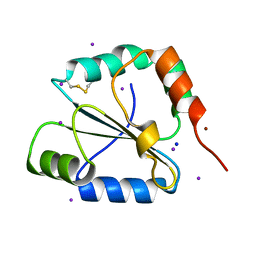 | | crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (oxidized form) | | Descriptor: | IODIDE ION, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
4V5M
 
 | | tRNA tranlocation on the 70S ribosome: the pre-translocational translocation intermediate TI(PRE) | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ratje, A.H, Loerke, J, Mikolajka, A, Bruenner, M, Hildebrand, P.W, Starosta, A.L, Doenhoefer, A, Connell, S.R, Fucini, P, Mielke, T, Whitford, P.C, Onuchic, J.N, Yu, Y, Sanbonmatsu, K.Y, Hartmann, R.K, Penczek, P.A, Wilson, D.N, Spahn, C.M.T. | | Deposit date: | 2010-10-01 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Head Swivel on the Ribosome Facilitates Translocation by Means of Intra-Subunit tRNA Hybrid Sites.
Nature, 468, 2010
|
|
2FWH
 
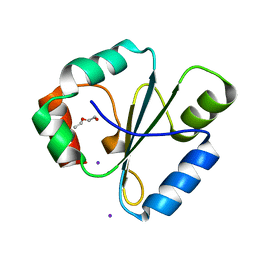 | | atomic resolution crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (reduced form at pH7) | | Descriptor: | DI(HYDROXYETHYL)ETHER, IODIDE ION, Thiol:disulfide interchange protein dsbD | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
2FWF
 
 | | high resolution crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (reduced form) | | Descriptor: | IODIDE ION, SODIUM ION, Thiol:disulfide interchange protein dsbD | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
2GO5
 
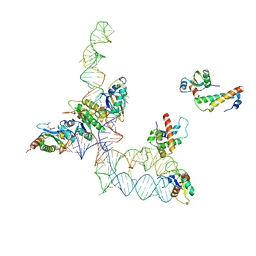 | | Structure of signal recognition particle receptor (SR) in complex with signal recognition particle (SRP) and ribosome nascent chain complex | | Descriptor: | SRP RNA, Signal recognition particle 19 kDa protein (SRP19), Signal recognition particle 54 kDa protein (SRP54), ... | | Authors: | Halic, M, Gartmann, M, Schlenker, O, Mielke, T, Pool, M.R, Sinning, I, Beckmann, R. | | Deposit date: | 2006-04-12 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Signal Recognition Particle Receptor Exposes the Ribosomal Translocon Binding Site
Science, 312, 2006
|
|
2MVF
 
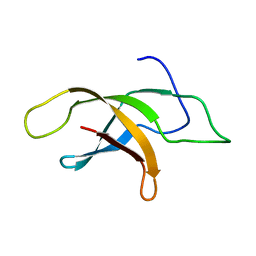 | | Structural insight into an essential assembly factor network on the pre-ribosome | | Descriptor: | Uncharacterized protein | | Authors: | Lee, W, Bassler, J, Paternoga, H, Holdermann, I, Thomas, M, Granneman, S, Barrio-Garcia, C, Nyarko, A, Stier, G, Clark, S.A, Schraivogel, D, Kallas, M, Beckmann, R, Tollervey, D, Barbar, E, Sinning, I, Hurt, E. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
4V7E
 
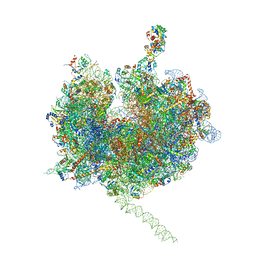 | | Model of the small subunit RNA based on a 5.5 A cryo-EM map of Triticum aestivum translating 80S ribosome | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S10E, ... | | Authors: | Barrio-Garcia, C, Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-11-22 | | Release date: | 2014-07-09 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion.
Nature, 506, 2014
|
|
4V6I
 
 | | Localization of the small subunit ribosomal proteins into a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein RACK1 (RACK1), ... | | Authors: | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2010-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V6W
 
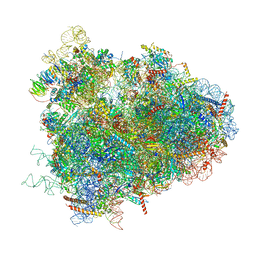 | | Structure of the D. melanogaster 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 2S ribosomal RNA, ... | | Authors: | Anger, A.M, Armache, J.-P, Berninghausen, O, Habeck, M, Subklewe, M, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-02-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structures of the human and Drosophila 80S ribosome.
Nature, 497, 2013
|
|
2KB6
 
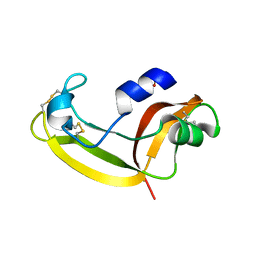 | | Solution structure of onconase C87A/C104A | | Descriptor: | Protein P-30 | | Authors: | Weininger, U, Schulenburg, C, Arnold, U, Ulbrich-Hofmann, R, Balbach, J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Impact of the C-terminal disulfide bond on the folding and stability of onconase.
Chembiochem, 11, 2010
|
|
4V6X
 
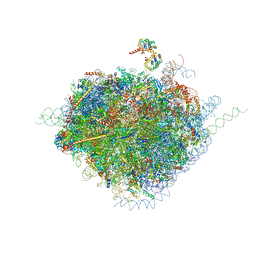 | | Structure of the human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Anger, A.M, Armache, J.-P, Berninghausen, O, Habeck, M, Subklewe, M, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-02-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structures of the human and Drosophila 80S ribosome.
Nature, 497, 2013
|
|
4UE5
 
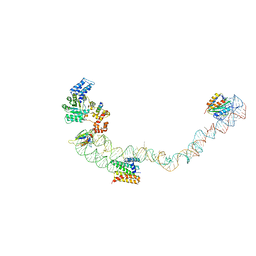 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | Descriptor: | 7S RNA, SIGNAL RECOGNITION PARTICLE 54 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, ... | | Authors: | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | Deposit date: | 2014-12-15 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UTQ
 
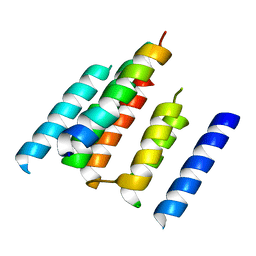 | | A structural model of the active ribosome-bound membrane protein insertase YidC | | Descriptor: | ATP SYNTHASE SUBUNIT C, MEMBRANE PROTEIN INSERTASE YIDC | | Authors: | Wickles, S, Singharoy, A, Andreani, J, Seemayer, S, Bischoff, L, Berninghausen, O, Soeding, J, Schulten, K, vanderSluis, E.O, Beckmann, R. | | Deposit date: | 2014-07-22 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A Structural Model of the Active Ribosome-Bound Membrane Protein Insertase Yidc.
Elife, 3, 2014
|
|
4UE4
 
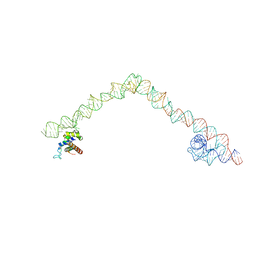 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | Descriptor: | 6S RNA, FTSQ SIGNAL SEQUENCE, SIGNAL RECOGNITION PARTICLE PROTEIN | | Authors: | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | Deposit date: | 2014-12-15 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7ABF
 
 | | Human pre-Bact-1 spliceosome core structure | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2020-12-09 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
4UY9
 
 | | Structure of MLK1 kinase domain with leucine zipper 1 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 9 | | Authors: | Read, J.A, Brassington, C, Pollard, H.K, Phillips, C, Green, I, Overmann, R, Collier, M. | | Deposit date: | 2014-08-29 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Recurrent Mlk4 Loss-of-Function Mutations Suppress Jnk Signaling to Promote Colon Tumorigenesis.
Cancer Res., 76, 2016
|
|
