1JA8
 
 | | Kinetic Analysis of Product Inhibition in Human Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, Manganese Superoxide Dismutase, SULFATE ION | | Authors: | Hearn, A.S, Stroupe, M.E, Cabelli, D.E, Lepock, J.R, Tainer, J.A, Nick, H.S, Silverman, D.S. | | Deposit date: | 2001-05-29 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Kinetic analysis of product inhibition in human manganese superoxide dismutase.
Biochemistry, 40, 2001
|
|
6B89
 
 | | E. coli LptB in complex with ADP and novobiocin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lipopolysaccharide export system ATP-binding protein LptB, MAGNESIUM ION, ... | | Authors: | May, J.M, Lazarus, M.B, Sherman, D.J, Owens, T.W, Mandler, M.D, Kahne, D.K. | | Deposit date: | 2017-10-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Antibiotic Novobiocin Binds and Activates the ATPase That Powers Lipopolysaccharide Transport.
J. Am. Chem. Soc., 139, 2017
|
|
7T39
 
 | | Co-crystal structure of human PRMT9 in complex with MT221 inhibitor | | Descriptor: | 7-[5-S-(4-{[(2-ethylpyridin-3-yl)methyl]amino}butyl)-5-thio-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Protein arginine N-methyltransferase 9 | | Authors: | Zeng, H, Dong, A, Hutchinson, A, Seitova, A, Li, Y, Gao, Y.D, Schneider, S, Siliphaivanh, P, Sloman, D, Nicholson, B, Fischer, C, Hicks, J, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-12-07 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Co-crystal structure of human PRMT9 in complex with MT221 inhibitor
To Be Published
|
|
7T3X
 
 | | Structure of unphosphorylated Pediculus humanus (Ph) PINK1 D334A mutant | | Descriptor: | Serine/threonine-protein kinase PINK1 | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
7T4K
 
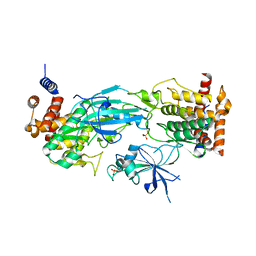 | | Structure of dimeric phosphorylated Pediculus humanus (Ph) PINK1 with kinked alpha-C helix in chain B | | Descriptor: | Serine/threonine-protein kinase PINK1, putative | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
7T4N
 
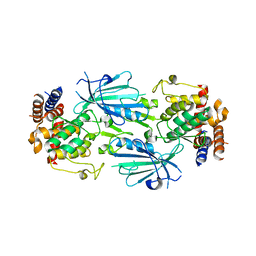 | | Structure of dimeric unphosphorylated Pediculus humanus (Ph) PINK1 D357A mutant | | Descriptor: | Serine/threonine-protein kinase PINK1, putative | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
7T4L
 
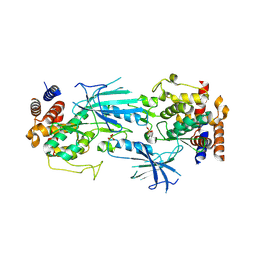 | | Structure of dimeric phosphorylated Pediculus humanus (Ph) PINK1 with extended alpha-C helix in chain B | | Descriptor: | Serine/threonine-protein kinase PINK1, putative | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
7T4M
 
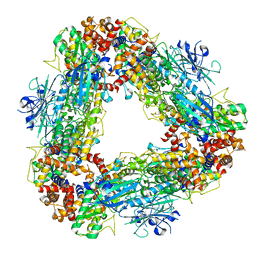 | | Structure of dodecameric unphosphorylated Pediculus humanus (Ph) PINK1 D357A mutant | | Descriptor: | Serine/threonine-protein kinase PINK1, putative | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
1JOC
 
 | | EEA1 homodimer of C-terminal FYVE domain bound to inositol 1,3-diphosphate | | Descriptor: | Early Endosomal Autoantigen 1, PHOSPHORIC ACID MONO-(2,3,4,6-TETRAHYDROXY-5-PHOSPHONOOXY-CYCLOHEXYL) ESTER, ZINC ION | | Authors: | Dumas, J.J, Merithew, E, Rajamani, D, Hayes, S, Lawe, D, Corvera, S, Lambright, D.G. | | Deposit date: | 2001-07-27 | | Release date: | 2001-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multivalent endosome targeting by homodimeric EEA1.
Mol.Cell, 8, 2001
|
|
7SWP
 
 | | G32Q4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | G32Q4 Fab heavy chain, G32Q4 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
4M11
 
 | | Crystal Structure of Murine Cyclooxygenase-2 Complex with Meloxicam | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-hydroxy-2-methyl-N-(5-methyl-1,3-thiazol-2-yl)-2H-1,2-benzothiazine-3-carboxamide 1,1-dioxide, ... | | Authors: | Xu, S, Banerjee, S, Hermanson, D.J, Marnett, L.J. | | Deposit date: | 2013-08-02 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Oxicams Bind in a Novel Mode to the Cyclooxygenase Active Site via a Two-water-mediated H-bonding Network.
J.Biol.Chem., 289, 2014
|
|
7TZJ
 
 | | SARS CoV-2 PLpro in complex with inhibitor 3k | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3-fluorophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Papain-like protease, ... | | Authors: | Calleja, D.J, Klemm, T, Lechtenberg, B.C, Kuchel, N.W, Lessene, G, Komander, D. | | Deposit date: | 2022-02-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Insights Into Drug Repurposing, as Well as Specificity and Compound Properties of Piperidine-Based SARS-CoV-2 PLpro Inhibitors.
Front Chem, 10, 2022
|
|
1KIB
 
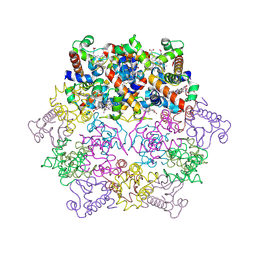 | | cytochrome c6 from Arthrospira maxima: an assembly of 24 subunits in the form of an oblate shell | | Descriptor: | HEME C, cytochrome c6 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Krogmann, D, Yeates, T.O. | | Deposit date: | 2001-12-03 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of cytochrome c6 from Arthrospira maxima: an assembly of 24 subunits in a nearly symmetric shell.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KAL
 
 | |
7TVG
 
 | |
7TVF
 
 | |
7S9Y
 
 | | Helicobacter Hepaticus CcsBA Open Conformation | | Descriptor: | Cytochrome c biogenesis protein, HEME B/C, PHOSPHATIDYLETHANOLAMINE | | Authors: | Mendez, D.L, Lowder, E.P, Tillman, D.E, Sutherland, M.C, Collier, A.L, Rau, M.J, Fitzpatrick, J.A, Kranz, R.G. | | Deposit date: | 2021-09-21 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM of CcsBA reveals the basis for cytochrome c biogenesis and heme transport.
Nat.Chem.Biol., 18, 2022
|
|
1JBD
 
 | | NMR Structure of the Complex Between alpha-bungarotoxin and a Mimotope of the Nicotinic Acetylcholine Receptor | | Descriptor: | LONG NEUROTOXIN 1, MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR | | Authors: | Scarselli, M, Spiga, O, Ciutti, A, Bracci, L, Lelli, B, Lozzi, L, Calamandrei, D, Bernini, A, Di Maro, D, Klein, S, Niccolai, N. | | Deposit date: | 2001-06-04 | | Release date: | 2001-06-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
4M10
 
 | | Crystal Structure of Murine Cyclooxygenase-2 Complex with Isoxicam | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-hydroxy-2-methyl-N-(5-methyl-1,2-oxazol-3-yl)-2H-1,2-benzothiazine-3-carboxamide 1,1-dioxide, ... | | Authors: | Xu, S, Hermanson, D.J, Banerjee, S, Ghebreelasie, K, Marnett, L.J. | | Deposit date: | 2013-08-02 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Oxicams Bind in a Novel Mode to the Cyclooxygenase Active Site via a Two-water-mediated H-bonding Network.
J.Biol.Chem., 289, 2014
|
|
1KEQ
 
 | | Crystal Structure of F65A/Y131C Carbonic Anhydrase V, covalently modified with 4-chloromethylimidazole | | Descriptor: | 4-METHYLIMIDAZOLE, ACETIC ACID, F65A/Y131C-MI Carbonic Anhydrase V, ... | | Authors: | Jude, K.M, Wright, S.K, Tu, C, Silverman, D.N, Viola, R.E, Christianson, D.W. | | Deposit date: | 2001-11-16 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of F65A/Y131C-methylimidazole carbonic anhydrase V reveals architectural features of an engineered proton shuttle.
Biochemistry, 41, 2002
|
|
1OP4
 
 | | Solution Structure of Neural Cadherin Prodomain | | Descriptor: | Neural-cadherin | | Authors: | Koch, A.W, Farooq, A, Shan, W, Zeng, L, Colman, D.R, Zhou, M.-M. | | Deposit date: | 2003-03-04 | | Release date: | 2004-03-16 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of the Neural (N-) Cadherin Prodomain Reveals a Cadherin Extracellular Domain-like Fold without Adhesive Characteristics
Structure, 12, 2004
|
|
1LKX
 
 | | MOTOR DOMAIN OF MYOE, A CLASS-I MYOSIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN IE HEAVY CHAIN, ... | | Authors: | Kollmar, M, Durrwang, U, Kliche, W, Manstein, D.J, Kull, F.J. | | Deposit date: | 2002-04-26 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the motor domain of a class-I myosin.
EMBO J., 21, 2002
|
|
1LUJ
 
 | | Crystal Structure of the Beta-catenin/ICAT Complex | | Descriptor: | Beta-catenin-interacting protein 1, Catenin beta-1 | | Authors: | Graham, T.A, Clements, W.K, Kimelman, D, Xu, W. | | Deposit date: | 2002-05-22 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the beta-catenin/ICAT complex reveals the inhibitory mechanism of ICAT.
Mol.Cell, 10, 2002
|
|
1LZV
 
 | | Site-Specific Mutant (Tyr7 replaced with His) of Human Carbonic Anhydrase II | | Descriptor: | Carbonic Anhydrase II, ZINC ION | | Authors: | Tu, C.K, Qian, M, An, H, Wadhwa, N.R, Duda, D.M, Yoshioka, C, Pathak, Y, McKenna, R, Laipis, P.J, Silverman, D.N. | | Deposit date: | 2002-06-11 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic analysis of multiple proton shuttles in the active site of human carbonic anhydrase.
J.Biol.Chem., 277, 2002
|
|
1LS1
 
 | |
