7MJL
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (focused refinement of RBD and Fab ab1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, Fab ab1 Light Chain, ... | | 著者 | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | 登録日 | 2021-04-20 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJN
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | 著者 | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | 登録日 | 2021-04-20 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJK
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (class 2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, ... | | 著者 | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | 登録日 | 2021-04-20 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJG
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | 登録日 | 2021-04-20 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJM
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | 登録日 | 2021-04-20 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
8UYF
 
 | | Structure of nucleotide-free Pediculus humanus (Ph) PINK1 dimer | | 分子名称: | Serine/threonine-protein kinase Pink1, mitochondrial | | 著者 | Gan, Z.Y, Kirk, N.S, Leis, A, Komander, D. | | 登録日 | 2023-11-13 | | 公開日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | Interaction of PINK1 with nucleotides and kinetin.
Sci Adv, 10, 2024
|
|
8W35
 
 | | Aca2 from Pectobacterium phage ZF40 bound to RNA | | 分子名称: | Anti-CRISPR associated (Aca) protein, Aca2, IR2 and IR-RBS RNA | | 著者 | Wilkinson, M.E, Birkholz, N, Kimanius, D, Fineran, P.C. | | 登録日 | 2024-02-21 | | 公開日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.61 Å) | | 主引用文献 | Phage anti-CRISPR control by an RNA- and DNA-binding helix-turn-helix protein.
Nature, 631, 2024
|
|
6BO7
 
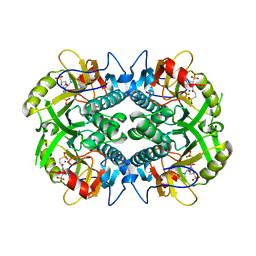 | | Crystal structure of Plasmodium vivax hypoxanthine guanine phosphoribosyltransferase in complex with [3R,4R]-4-guanin-9-yl-3-((S)-2-hydroxy-2-phosphonoethyl)oxy-1-N-(phosphonopropionyl)pyrrolidine | | 分子名称: | Hypoxanthine phosphoribosyltransferase, MAGNESIUM ION, [3-[(3~{R},4~{R})-3-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-[(2~{S})-2-oxidanyl-2-phosphono-ethoxy]pyrrolidin-1-y l]-3-oxidanylidene-propyl]phosphonic acid | | 著者 | Guddat, L.W, Keough, D.T, Rejman, D. | | 登録日 | 2017-11-18 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.856 Å) | | 主引用文献 | Design of Plasmodium vivax Hypoxanthine-Guanine Phosphoribosyltransferase Inhibitors as Potential Antimalarial Therapeutics.
ACS Chem. Biol., 13, 2018
|
|
3VVV
 
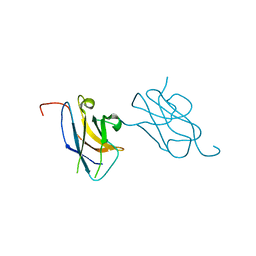 | | Skich domain of NDP52 | | 分子名称: | Calcium-binding and coiled-coil domain-containing protein 2 | | 著者 | Akutsu, M, Muhlinen, N.V, Randow, F, Komander, D. | | 登録日 | 2012-07-28 | | 公開日 | 2013-02-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy
Mol.Cell, 48, 2012
|
|
5UJL
 
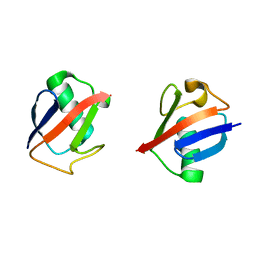 | |
5UJN
 
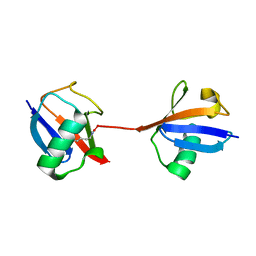 | |
5WPS
 
 | | Crystal structure HpiC1 Y101F | | 分子名称: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | 登録日 | 2017-08-07 | | 公開日 | 2018-04-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.389 Å) | | 主引用文献 | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
6CZ7
 
 | | The arsenate respiratory reductase (Arr) complex from Shewanella sp. ANA-3 | | 分子名称: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4Fe-4S ferredoxin, ... | | 著者 | Glasser, N.R, Newman, D.K. | | 登録日 | 2018-04-08 | | 公開日 | 2018-08-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Structural and mechanistic analysis of the arsenate respiratory reductase provides insight into environmental arsenic transformations.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CZ9
 
 | | The arsenate respiratory reductase (Arr) complex from Shewanella sp. ANA-3 bound to arsenite | | 分子名称: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4Fe-4S ferredoxin, ... | | 著者 | Glasser, N.R, Newman, D.K. | | 登録日 | 2018-04-08 | | 公開日 | 2018-08-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and mechanistic analysis of the arsenate respiratory reductase provides insight into environmental arsenic transformations.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CZ8
 
 | | The arsenate respiratory reductase (Arr) complex from Shewanella sp. ANA-3 bound to arsenate | | 分子名称: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4Fe-4S ferredoxin, ... | | 著者 | Glasser, N.R, Newman, D.K. | | 登録日 | 2018-04-08 | | 公開日 | 2018-08-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structural and mechanistic analysis of the arsenate respiratory reductase provides insight into environmental arsenic transformations.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CZA
 
 | | The arsenate respiratory reductase (Arr) complex from Shewanella sp. ANA-3 bound to phosphate | | 分子名称: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4Fe-4S ferredoxin, ... | | 著者 | Glasser, N.R, Newman, D.K. | | 登録日 | 2018-04-08 | | 公開日 | 2018-08-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structural and mechanistic analysis of the arsenate respiratory reductase provides insight into environmental arsenic transformations.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6D71
 
 | | Crystal Structure of the Human Miro1 N-terminal GTPase bound to GTP | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial Rho GTPase 1 | | 著者 | Smith, K.P, Focia, P.J, Rice, S.E, Freymann, D.M. | | 登録日 | 2018-04-23 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.7180779 Å) | | 主引用文献 | Insight into human Miro1/2 domain organization based on the structure of its N-terminal GTPase.
J.Struct.Biol., 212, 2020
|
|
6DXK
 
 | | Glucocorticoid Receptor in complex with Compound 11 | | 分子名称: | (8S,11R,13S,14S,17S)-11-[4-(dimethylamino)phenyl]-17-(3,3-dimethylbut-1-yn-1-yl)-17-hydroxy-13-methyl-1,2,6,7,8,11,12,13,14,15,16,17-dodecahydro-3H-cyclopenta[a]phenanthren-3-one (non-preferred name), Glucocorticoid receptor | | 著者 | Rew, Y, Du, X, Eksterowicz, J, Zhou, H, Jahchan, N, Zhu, L, Yan, X, Kawai, H, McGee, L.R, Medina, J.C, Huang, T, Chen, C, Zavorotinskaya, T, Sutimantanapi, D, Waszczuk, J, Jackson, E, Huang, E, Ye, Q, Fantin, V.R, Daqing, S. | | 登録日 | 2018-06-29 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Discovery of a Potent and Selective Steroidal Glucocorticoid Receptor Antagonist (ORIC-101).
J. Med. Chem., 61, 2018
|
|
5RL3
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-39 (Mpro-x3117) | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-tert-butylphenyl)-N-[(1R)-2-[(oxan-4-yl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-08-05 | | 公開日 | 2020-12-02 | | 最終更新日 | 2021-07-07 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
5RL2
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-26 (Mpro-x3115) | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-tert-butylphenyl)-N-[(1R)-2-[(2-methoxyethyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-08-05 | | 公開日 | 2020-12-02 | | 最終更新日 | 2021-07-07 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
5RL0
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-2 (Mpro-x3110) | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ethyl N-[(2R)-2-[(4-tert-butylphenyl)(propanoyl)amino]-2-(pyridin-3-yl)acetyl]-beta-alaninate | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-08-05 | | 公開日 | 2020-12-02 | | 最終更新日 | 2021-07-07 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
5RL1
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-27 (Mpro-x3113) | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-tert-butylphenyl)-N-[(1R)-2-[(3-methoxypropyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-08-05 | | 公開日 | 2020-12-02 | | 最終更新日 | 2021-07-07 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
5RL5
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-30 (Mpro-x3359) | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-tert-butylphenyl)-N-[(1R)-2-(ethylamino)-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-08-05 | | 公開日 | 2020-12-02 | | 最終更新日 | 2021-07-07 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
6CU6
 
 | |
4BS9
 
 | | Structure of the heterocyclase TruD | | 分子名称: | TRUD, ZINC ION | | 著者 | Koehnke, J, Bent, A.F, Zollman, D, Smith, K, Houssen, W.E, Zhu, X, Mann, G, Lebl, T, Scharff, R, Shirran, S, Botting, C.H, Jaspars, M, Schwarz-Linek, U, Naismith, J.H. | | 登録日 | 2013-06-09 | | 公開日 | 2013-11-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The Cyanobactin Heterocyclase Enzyme: A Processive Adenylase that Operates with a Defined Order of Reaction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
