6LLQ
 
 | | Solution NMR structure of de novo Rossmann2x2 fold with most of the core mutated to valine, R2x2_VAL88 | | Descriptor: | VAL88 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Koga, R, Yamamoto, M, Kosugi, T, Koga, N. | | Deposit date: | 2019-12-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Robust folding of a de novo designed ideal protein even with most of the core mutated to valine.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5Y5G
 
 | | Structure of cytochrome P450nor in NO-bound state: damaged by high-dose (5.7 MGy) X-ray | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
1GC0
 
 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT L-METHIONINE GAMMA-LYASE FROM PSEUDOMONAS PUTIDA | | Descriptor: | METHIONINE GAMMA-LYASE | | Authors: | Motoshima, H, Inagaki, K, Kumasaka, T, Furuichi, M, Inoue, H, Tamura, T, Esaki, N, Soda, K, Tanaka, N, Yamamoto, M, Tanaka, H. | | Deposit date: | 2000-07-06 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J.Biochem., 128, 2000
|
|
7DVO
 
 | | Structure of Reaction Intermediate of Cytochrome P450 NO Reductase (P450nor) Determined by XFEL | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Nomura, T, Kimura, T, Kanematsu, Y, Yamashita, K, Hirata, K, Ueno, G, Murakami, H, Hisano, T, Yamagiwa, R, Takeda, H, Gopalasingam, C, Yuki, K, Kousaka, R, Yanagasawa, S, Shoji, O, Kumasaka, T, Takano, Y, Ago, H, Yamamoto, M, Sugimoto, H, Tosha, T, Kubo, M, Shiro, Y. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Short-lived intermediate in N 2 O generation by P450 NO reductase captured by time-resolved IR spectroscopy and XFEL crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5XFE
 
 | | Luciferin-regenerating enzyme solved by SAD using XFEL (refined against 11,000 patterns) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2017-04-10 | | Release date: | 2017-08-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental phase determination with selenomethionine or mercury-derivatization in serial femtosecond crystallography
IUCrJ, 4, 2017
|
|
1V3V
 
 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase complexed with NADP and 15-oxo-PGE2 | | Descriptor: | (5E,13E)-11-HYDROXY-9,15-DIOXOPROSTA-5,13-DIEN-1-OIC ACID, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-06 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
5H6K
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 | | Descriptor: | 1,2-ETHANEDIOL, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5D9B
 
 | | Luciferin-regenerating enzyme solved by SIRAS using XFEL (refined against native data) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
5H6N
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1, autoinhibitory form | | Descriptor: | Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
1V3T
 
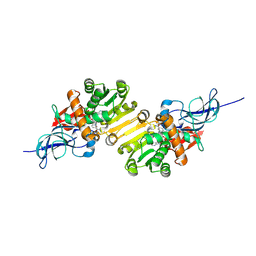 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, leukotriene b4 12-hydroxydehydrogenase/prostaglandin 15-keto reductase | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-05 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
5H6L
 
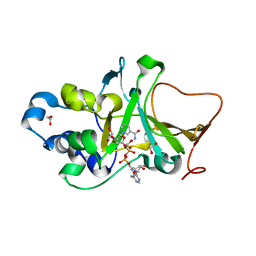 | | DNA targeting ADP-ribosyltransferase Pierisin-1 in complex with beta-NAD+ | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6M
 
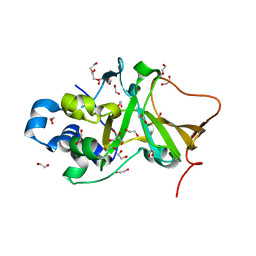 | | DNA targeting ADP-ribosyltransferase Pierisin-1 | | Descriptor: | 1,2-ETHANEDIOL, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
1V26
 
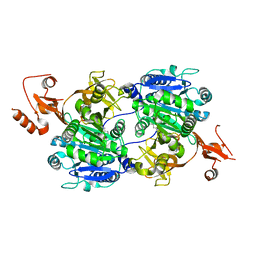 | | Crystal structure of tt0168 from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, MYRISTIC ACID, ... | | Authors: | Hisanaga, Y, Ago, H, Nakatsu, T, Hamada, K, Ida, K, Kanda, H, Yamamoto, M, Hori, T, Arii, Y, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-07 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of the Substrate-specific Two-step Catalysis of Long Chain Fatty Acyl-CoA Synthetase Dimer
J.Biol.Chem., 279, 2004
|
|
1ULU
 
 | | Crystal structure of tt0143 from Thermus thermophilus HB8 | | Descriptor: | enoyl-acyl carrier protein reductase | | Authors: | Ago, H, Hamada, K, Ida, K, Kanda, H, Sugahara, M, Yamamoto, M, Nodake, Y, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of tt0143 from Thermus thermophilus HB8
To be Published
|
|
1ULT
 
 | | Crystal structure of tt0168 from Thermus thermophilus HB8 | | Descriptor: | CITRIC ACID, long chain fatty acid-CoA ligase | | Authors: | Hisanaga, Y, Ago, H, Nakatsu, T, Hamada, K, Ida, K, Kanda, H, Yamamoto, M, Hori, T, Arii, Y, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of the substrate specific two-step catalysis of long chain fatty acyl-CoA synthetase dimer
J.Biol.Chem., 279, 2004
|
|
5D9D
 
 | | Luciferin-regenerating enzyme solved by SAD using synchrotron radiation at room temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
1ULQ
 
 | | Crystal structure of tt0182 from Thermus thermophilus HB8 | | Descriptor: | putative acetyl-CoA acetyltransferase | | Authors: | Ago, H, Hamada, K, Ida, K, Kanda, H, Sugahara, M, Yamamoto, M, Kuroishi, C, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of tt0182 from Thermus thermophilus HB8
To be Published
|
|
5D9C
 
 | | Luciferin-regenerating enzyme solved by SIRAS using XFEL (refined against Hg derivative data) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
1V25
 
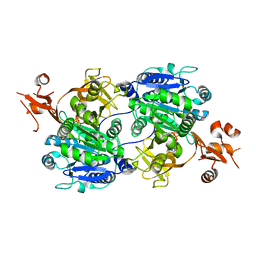 | | Crystal structure of tt0168 from Thermus thermophilus HB8 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, long-chain-fatty-acid-CoA synthetase | | Authors: | Hisanaga, Y, Ago, H, Nakatsu, T, Hamada, K, Ida, K, Kanda, H, Yamamoto, M, Hori, T, Arii, Y, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-07 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Substrate-specific Two-step Catalysis of Long Chain Fatty Acyl-CoA Synthetase Dimer
J.Biol.Chem., 279, 2004
|
|
1V3U
 
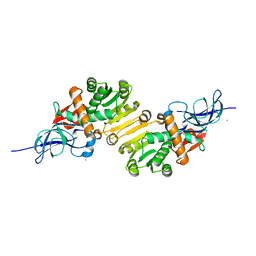 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase in apo form | | Descriptor: | CHLORIDE ION, leukotriene b4 12-hydroxydehydrogenase/prostaglandin 15-keto reductase | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-05 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
1IOO
 
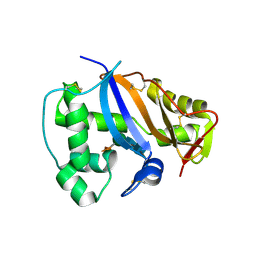 | | CRYSTAL STRUCTURE OF NICOTIANA ALATA GEMETOPHYTIC SELF-INCOMPATIBILITY ASSOCIATED SF11-RNASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-xylopyranose-(1-2)][alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SF11-RNASE, beta-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ida, K, Sato, M, Sakiyama, F, Norioka, S, Yamamoto, M, Kumasaka, T, Yamashita, E. | | Deposit date: | 2001-03-26 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The 1.55 A resolution structure of Nicotiana alata S(F11)-RNase associated with gametophytic self-incompatibility.
J.Mol.Biol., 314, 2001
|
|
6I5C
 
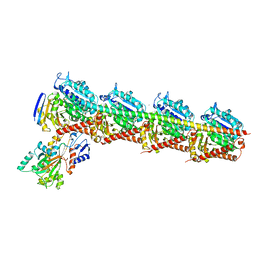 | | Long wavelength native-SAD phasing of Tubulin-Stathmin-TTL complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Basu, S, Olieric, V, Matsugaki, N, Kawano, Y, Takashi, T, Huang, C.Y, Leonarski, F, Yamada, Y, Vera, L, Olieric, N, Basquin, J, Wojdyla, J.A, Diederichs, K, Yamamoto, M, Bunk, O, Wang, M. | | Deposit date: | 2018-11-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Long-wavelength native-SAD phasing: opportunities and challenges.
Iucrj, 6, 2019
|
|
1WKY
 
 | | Crystal structure of alkaline mannanase from Bacillus sp. strain JAMB-602: catalytic domain and its Carbohydrate Binding Module | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Akita, M, Takeda, N, Hirasawa, K, Sakai, H, Kawamoto, M, Yamamoto, M, Grant, W.D, Hatada, Y, Ito, S, Horikoshi, K. | | Deposit date: | 2004-06-15 | | Release date: | 2005-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallization and preliminary X-ray study of alkaline mannanase from an alkaliphilic Bacillus isolate.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1X2J
 
 | | Structural basis for the defects of human lung cancer somatic mutations in the repression activity of Keap1 on Nrf2 | | Descriptor: | Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Padmanabhan, B, Tong, K.I, Nakamura, Y, Ohta, T, Scharlock, M, Kobayashi, A, Ohtsuji, M, Kang, M.-I, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-04-25 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for defects of keap1 activity provoked by its point mutations in lung cancer
Mol.Cell, 21, 2006
|
|
1X2R
 
 | | Structural basis for the defects of human lung cancer somatic mutations in the repression activity of Keap1 on Nrf2 | | Descriptor: | Kelch-like ECH-associated protein 1, Nuclear factor erythroid 2 related factor 2, SULFATE ION | | Authors: | Padmanabhan, B, Tong, K.I, Nakamura, Y, Ohta, T, Scharlock, M, Kobayashi, A, Ohtsuji, M, Kang, M.-I, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-04-26 | | Release date: | 2006-03-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for defects of keap1 activity provoked by its point mutations in lung cancer
Mol.Cell, 21, 2006
|
|
