4M9Q
 
 | | Crystal structure of C-terminally truncated Arl13B from Chlamydomonas rheinhardtii bound to GppNHp | | Descriptor: | ARF-like GTPase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Miertzschke, M, Koerner, C, Spoerner, M, Wittinghofer, A. | | Deposit date: | 2013-08-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the small G-protein Arl13B and implications for Joubert syndrome.
Biochem.J., 457, 2014
|
|
1UA7
 
 | | Crystal Structure Analysis of Alpha-Amylase from Bacillus Subtilis complexed with Acarbose | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha-amylase, ... | | Authors: | Kagawa, M, Fujimoto, Z, Momma, M, Takase, K, Mizuno, H. | | Deposit date: | 2003-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of Bacillus subtilis alpha-amylase in complex with acarbose
J.BACTERIOL., 185, 2003
|
|
6RNK
 
 | | Crystal structure of a humanized (K18E, K269N) rat succinate receptor SUCNR1 (GPR91) in complex with a nanobody and antagonist NF-56-EJ40. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[2-[[3-[4-[(4-methylpiperazin-1-yl)methyl]phenyl]phenyl]carbonylamino]phenyl]ethanoic acid, GLYCEROL, ... | | Authors: | Haffke, M, Jaakola, V.-P. | | Deposit date: | 2019-05-08 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis of species-selective antagonist binding to the succinate receptor.
Nature, 574, 2019
|
|
1IT5
 
 | | Solution structure of apo-type PLA2 from Streptomyces violaceruber A-2688. | | Descriptor: | Phospholipase A2 | | Authors: | Sugiyama, M, Ohtani, K, Izuhara, M, Koike, T. | | Deposit date: | 2002-01-09 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel prokaryotic phospholipase A2. Characterization, gene cloning, and solution structure.
J.Biol.Chem., 277, 2002
|
|
2MK1
 
 | |
8EVF
 
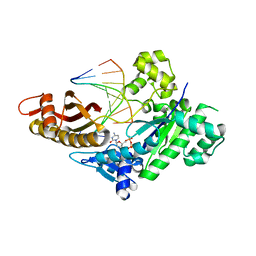 | | HUMAN DNA POLYMERASE ETA EXTENSION COMPLEX WITH AN INCOMING DCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*G)-3'), ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2022-10-20 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | The peroxidation-derived DNA adduct, 6-oxo-M 1 dG, is a strong block to replication by human DNA polymerase eta.
J.Biol.Chem., 299, 2023
|
|
8EVE
 
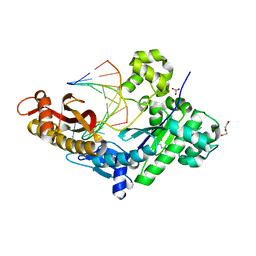 | | HUMAN DNA POLYMERASE ETA INSERTION COMPLEX | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2022-10-20 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The peroxidation-derived DNA adduct, 6-oxo-M 1 dG, is a strong block to replication by human DNA polymerase eta.
J.Biol.Chem., 299, 2023
|
|
7E57
 
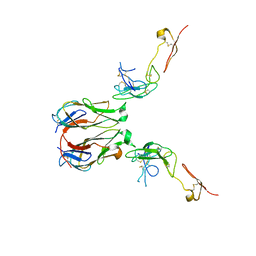 | | Crystal structure of murine GITR-GITRL complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 18, Tumor necrosis factor receptor superfamily member 18, ... | | Authors: | Zhao, M, Tan, S, Fu, L, Chai, Y, Qi, J, Gao, G.F. | | Deposit date: | 2021-02-18 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Atypical TNF-TNFR superfamily binding interface in the GITR-GITRL complex for T cell activation.
Cell Rep, 36, 2021
|
|
1UGN
 
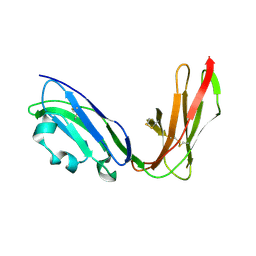 | | Crystal structure of LIR1.02, one of the alleles of LIR1 | | Descriptor: | Leukocyte immunoglobulin-like receptor 1 | | Authors: | Shiroishi, M, Rasubala, L, Kuroki, K, Amano, K, Tsuchiya, N, Tokunaga, K, Kohda, D, Maenaka, K. | | Deposit date: | 2003-06-17 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Extensive polymorphisms of LILRB1 (ILT2, LIR1) and their association with HLA-DRB1 shared epitope negative rheumatoid arthritis.
Hum.Mol.Genet., 14, 2005
|
|
5A0E
 
 | | Crystal structure of cyclophilin D in complex with CsA analogue, JW47. | | Descriptor: | JW47, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE F, MITOCHONDRIAL | | Authors: | Warne, J, Pryce, G, Hill, J, Shi, X, Lenneras, F, Puentes, F, Kip, M, Hilditch, L, Walker, P, Simone, M, Chan, A.W.E, Towers, G, Coker, A.R, Duchen, M, Szabadkai, G, Baker, D, Selwood, D.L. | | Deposit date: | 2015-04-19 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Selective Inhibition of the Mitochondrial Permeability Transition Pore Protects Against Neuro-Degeneration in Experimental Multiple Sclerosis.
J.Biol.Chem., 291, 2016
|
|
7ZEI
 
 | | Thermostable GH159 glycoside hydrolase from Caldicellulosiruptor at 1.7 A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Baudrexl, M, Fida, T, Berk, B, Schwarz, W, Zverlov, V.V, Groll, M, Liebl, W. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and Structural Characterization of Thermostable GH159 Glycoside Hydrolases Exhibiting alpha-L-Arabinofuranosidase Activity.
Front Mol Biosci, 9, 2022
|
|
6YEJ
 
 | | Cryo-EM structure of the Full-length disease type human Huntingtin | | Descriptor: | Huntingtin | | Authors: | Tame, G, Jung, T, Dal Perraro, M, Hebert, H, Song, J. | | Deposit date: | 2020-03-24 | | Release date: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (18.200001 Å) | | Cite: | The Polyglutamine Expansion at the N-Terminal of Huntingtin Protein Modulates the Dynamic Configuration and Phosphorylation of the C-Terminal HEAT Domain.
Structure, 28, 2020
|
|
1ION
 
 | | THE SEPTUM SITE-DETERMINING PROTEIN MIND COMPLEXED WITH MG-ADP FROM PYROCOCCUS HORIKOSHII OT3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROBABLE CELL DIVISION INHIBITOR MIND | | Authors: | Sakai, N, Yao, M, Itou, H, Watanabe, N, Yumoto, F, Tanokura, M, Tanaka, I. | | Deposit date: | 2001-03-21 | | Release date: | 2001-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of septum site-determining protein MinD from Pyrococcus horikoshii OT3 in complex with Mg-ADP.
Structure, 9, 2001
|
|
1IPK
 
 | | CRYSTAL STRUCTURES OF RECOMBINANT AND NATIVE SOYBEAN BETA-CONGLYCININ BETA HOMOTRIMERS | | Descriptor: | BETA-CONGLYCININ, BETA CHAIN | | Authors: | Maruyama, N, Adachi, M, Takahashi, K, Yagasaki, K, Kohno, M, Takenaka, Y, Okuda, E, Nakagawa, S, Mikami, B, Utsumi, S. | | Deposit date: | 2001-05-16 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of recombinant and native soybean beta-conglycinin beta homotrimers.
Eur.J.Biochem., 268, 2001
|
|
1IQV
 
 | |
4ZSD
 
 | | Human Cyclophilin D Complexed with an Inhibitor at room temperature | | Descriptor: | 1-(4-aminobenzyl)-3-[(2S)-4-(methylsulfanyl)-1-{(2R)-2-[2-(methylsulfanyl)phenyl]pyrrolidin-1-yl}-1-oxobutan-2-yl]urea, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Gelin, M, Delfosse, V, Allemand, F, Hoh, F, Sallaz-Damaz, Y, Pirocchi, M, Bourguet, W, Ferrer, J.-L, Labesse, G, Guichou, J.-F. | | Deposit date: | 2015-05-13 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8ERC
 
 | |
1IVU
 
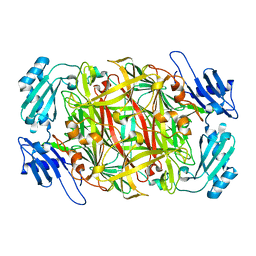 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Initial intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
6Y9P
 
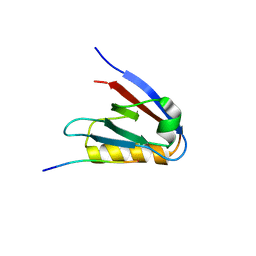 | | Crystal structure of Whirlin PDZ3_C-ter in complex with Harmonin a1 C-terminal PDZ binding motif peptide | | Descriptor: | Harmonin a1, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-03-10 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.169 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
6IPV
 
 | |
7EA6
 
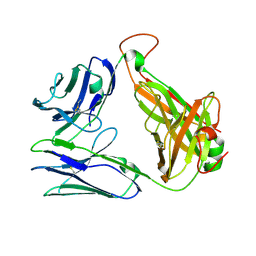 | | Crystal structure of TCR-017 ectodomain | | Descriptor: | T cell receptor 017 alpha chain, T cell receptor 017 beta chain | | Authors: | Nagae, M, Yamasaki, S. | | Deposit date: | 2021-03-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18000245 Å) | | Cite: | Identification of conserved SARS-CoV-2 spike epitopes that expand public cTfh clonotypes in mild COVID-19 patients.
J.Exp.Med., 218, 2021
|
|
5A7Y
 
 | | Crystal structure of Sulfolobus acidocaldarius Trm10 in complex with S-adenosylhomocysteine | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Van Laer, B, Roovers, M, Wauters, L, Kasprzak, J, Dyzma, M, Deyaert, E, Feller, A, Bujnicki, J, Droogmans, L, Versees, W. | | Deposit date: | 2015-07-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Insights Into tRNA Binding and Adenosine N1-Methylation by an Archaeal Trm10 Homologue.
Nucleic Acids Res., 44, 2016
|
|
5Y1B
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with a berberine derivative (SYSU-00679) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-3'-quinolinium propylberberine, Beta-hexosaminidase | | Authors: | Duan, Y.W, Liu, T, Zhou, Y, Tang, J.Y, Li, M, Yang, Q. | | Deposit date: | 2017-07-20 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.207 Å) | | Cite: | Glycoside hydrolase family 18 and 20 enzymes are novel targets of the traditional medicine berberine.
J. Biol. Chem., 293, 2018
|
|
6RXL
 
 | | Crystal structure of CobB wt in complex with H4K16-Crotonyl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1J03
 
 | | Solution structure of a putative steroid-binding protein from Arabidopsis | | Descriptor: | putative steroid binding protein | | Authors: | Suzuki, S, Hatanaka, H, Kigawa, T, Terada, T, Shirouzu, M, Seki, M, Shinozaki, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-29 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an Arabidopsis homologue of the mammalian membrane-associated progesterone receptor
To be Published
|
|
