4GNR
 
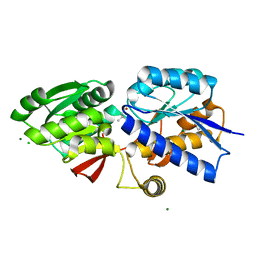 | | 1.0 Angstrom resolution crystal structure of the branched-chain amino acid transporter substrate binding protein LivJ from Streptococcus pneumoniae str. Canada MDR_19A in complex with Isoleucine | | Descriptor: | ABC transporter substrate-binding protein-branched chain amino acid transport, CHLORIDE ION, ISOLEUCINE, ... | | Authors: | Halavaty, A.S, Kudritska, M, Wawrzak, Z, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-17 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | 1.0 Angstrom resolution crystal structure of the branched-chain amino acid transporter substrate binding protein LivJ from Streptococcus pneumoniae str. Canada MDR_19A in complex with Isoleucine
To be Published
|
|
2XAN
 
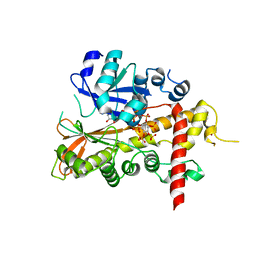 | | inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with AMP PNP and IP5 | | Descriptor: | INOSITOL-PENTAKISPHOSPHATE 2-KINASE, MAGNESIUM ION, MYO-INOSITOL-(1,3,4,5,6)-PENTAKISPHOSPHATE, ... | | Authors: | Gonzalez, B, Banos-Sanz, J.I, Villate, M, Brearley, C.A, Sanz-Aparicio, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase is a Distant Ipk Member with a Singular Inositide Binding Site for Axial 2-Oh Recognition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1XTZ
 
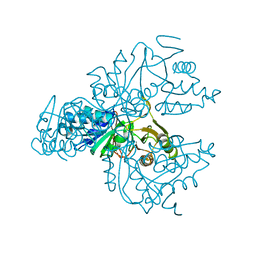 | | Crystal structure of the S. cerevisiae D-ribose-5-phosphate isomerase: comparison with the archeal and bacterial enzymes | | Descriptor: | Ribose-5-phosphate isomerase | | Authors: | Graille, M, Meyer, P, Leulliot, N, Sorel, I, Janin, J, van Tilbeurgh, H, Quevillon-Cheruel, S. | | Deposit date: | 2004-10-25 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the S. cerevisiae D-ribose-5-phosphate isomerase: comparison with the archaeal and bacterial enzymes
Biochimie, 87, 2005
|
|
2WT7
 
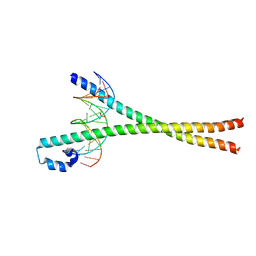 | | Crystal structure of the bZIP heterodimeric complex MafB:cFos bound to DNA | | Descriptor: | MODIFIED T-MARE MOTIF, PHOSPHATE ION, PROTO-ONCOGENE PROTEIN C-FOS, ... | | Authors: | Pogenberg, V, Holton, S, Wilmanns, M. | | Deposit date: | 2009-09-11 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of a bZIP Transcription Factor with Homo/Heterodimer-Induced DNA-Binding Preference.
Structure, 22, 2014
|
|
8B6R
 
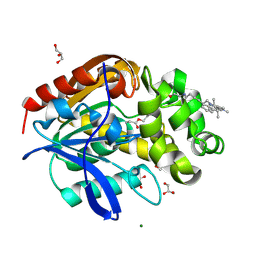 | | X-ray structure of the haloalkane dehalogenase HaloTag7 labeled with a chloroalkane Cyanine3 fluorophore substrate | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Hellweg, L, Hiblot, J. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A general method for the development of multicolor biosensors with large dynamic ranges.
Nat.Chem.Biol., 19, 2023
|
|
3M4D
 
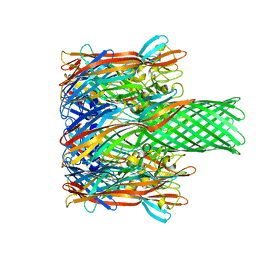 | |
1XU6
 
 | | Structure of the C-terminal domain from Trypanosoma brucei Variant Surface Glycoprotein MITat1.2 | | Descriptor: | Variant surface glycoprotein MITAT 1.2 | | Authors: | Chattopadhyay, A, Jones, N.G, Nietlispach, D, Nielsen, P.R, Voorheis, H.P, Mott, H.R, Carrington, M. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-30 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal domain from Trypanosoma brucei variant surface glycoprotein MITat1.2
J.Biol.Chem., 280, 2004
|
|
8BHW
 
 | |
8B42
 
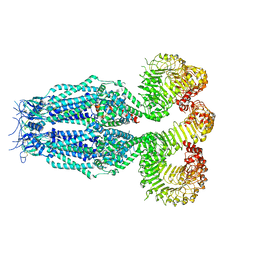 | |
5A95
 
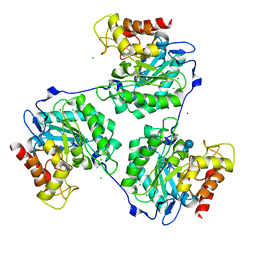 | | Crystal structure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-17 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
2WXV
 
 | | Structure of CDK2-CYCLIN A with a Pyrazolo(4,3-h) quinazoline-3- carboxamide inhibitor | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, N,1-DIMETHYL-8-{[1-(METHYLSULFONYL)PIPERIDIN-4-YL]AMINO}-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE, ... | | Authors: | Traquandi, G, Ciomei, M, Ballinari, D, Casale, E, Colombo, N, Croci, V, Fiorentini, F, Isacchi, A, Longo, A, Mercurio, C, Panzeri, A, Pastori, W, Pevarello, P, Volpi, D, Roussel, P, Vulpetti, A, Brasca, M.G. | | Deposit date: | 2009-11-10 | | Release date: | 2010-02-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Potent Pyrazolo[4,3-H]Quinazoline-3-Carboxamides as Multi-Cyclin-Dependent Kinase Inhibitors.
J.Med.Chem., 53, 2010
|
|
4GPX
 
 | | Crystal structure of the protozoal cytoplasmic ribosomal decoding site in complex with 6'-hydroxysisomicin (P212121 form) | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(hydroxymethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Koganei, M, Maianti, J.P, Ly, V.L, Hanessian, S. | | Deposit date: | 2012-08-22 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of a bioactive 6'-hydroxy variant of sisomicin bound to the bacterial and protozoal ribosomal decoding sites
Chemmedchem, 8, 2013
|
|
3M61
 
 | | Crystal structure of complex of urokinase and a upain-1 variant(W3A) in pH4.6 condition | | Descriptor: | Urokinase-type plasminogen activator, upain-1 W3A | | Authors: | Jiang, L, Yuan, C, Wind, T, Andreasen, P.A, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of complex of urokinase and a upain-1 variant(W3A) in pH4.6 condition
TO BE PUBLISHED
|
|
3M6E
 
 | |
3M2V
 
 | | Structural Insight into Methyl-Coenzyme M Reductase Chemistry using Coenzyme B Analogues | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Cedervall, P.E, Dey, M, Ragsdale, S.W, Wilmot, C.M. | | Deposit date: | 2010-03-08 | | Release date: | 2010-09-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.
Biochemistry, 49, 2010
|
|
8B1A
 
 | | DtpB-Nb132-AI | | Descriptor: | ALA-ILE, DECANE, DODECANE, ... | | Authors: | Killer, M, Finocchio, G, Lei, J, Jungnickel, K, Kotov, V, Steinke, J, Bartels, K, Strauss, J, Dupeux, F, Humm, A.S, Cornaciu, I, Marquez, J, Pardon, E, Steyeart, J, Loew, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Plasticity of the binding pocket in peptide transporters underpins promiscuous substrate recognition.
Cell Rep, 42, 2023
|
|
4GZB
 
 | | Crystal structure of native AmpC beta-lactamase from Pseudomonas aeruginosa PAO1 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Benvenuti, M, De Luca, F, Docquier, J.D, Mangani, S. | | Deposit date: | 2012-09-06 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural insight into potent broad-spectrum inhibition with reversible recyclization mechanism: avibactam in complex with CTX-M-15 and Pseudomonas aeruginosa AmpC beta-lactamases
Antimicrob.Agents Chemother., 57, 2013
|
|
8B1E
 
 | | DtpB-Nb132-AQ | | Descriptor: | ALA-GLN, DECANE, DODECANE, ... | | Authors: | Killer, M, Finocchio, G, Lei, J, Jungnickel, K, Kotov, V, Steinke, J, Bartels, K, Strauss, J, Dupeux, F, Humm, A.S, Cornaciu, I, Marquez, J, Pardon, E, Steyeart, J, Loew, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Plasticity of the binding pocket in peptide transporters underpins promiscuous substrate recognition.
Cell Rep, 42, 2023
|
|
2WX2
 
 | | X-RAY STRUCTURE OF CYP51 FROM THE HUMAN PATHOGEN TRYPANOSOMA CRUZI IN COMPLEX WITH FLUCONAZOLE | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, LANOSTEROL 14-ALPHA-DEMETHYLASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chen, C.-K, Leung, S.S.F, Guilbert, C, Jacobson, M, McKerrow, J.H, Podust, L.M. | | Deposit date: | 2009-10-31 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Characterization of Cyp51 from Trypanosoma Cruzi and Trypanosoma Brucei Bound to the Antifungal Drugs Posaconazole and Fluconazole
Plos Negl Trop Dis, 4, 2010
|
|
3M4G
 
 | | H57A HFQ from Pseudomonas Aeruginosa | | Descriptor: | Protein hfq, ZINC ION | | Authors: | Moskaleva, O, Melnik, B, Gabdulkhakov, A, Garber, M, Nikonov, S, Stolboushkina, E, Nikulin, A. | | Deposit date: | 2010-03-11 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structures of mutant forms of Hfq from Pseudomonas aeruginosa reveal the importance of the conserved His57 for the protein hexamer organization.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
8B1G
 
 | | DtpB-Nb132-AW | | Descriptor: | ALA-TRP, DECANE, DODECANE, ... | | Authors: | Killer, M, Finocchio, G, Lei, J, Jungnickel, K, Kotov, V, Steinke, J, Bartels, K, Strauss, J, Dupeux, F, Humm, A.S, Cornaciu, I, Marquez, J, Pardon, E, Steyeart, J, Loew, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Plasticity of the binding pocket in peptide transporters underpins promiscuous substrate recognition.
Cell Rep, 42, 2023
|
|
3M4Z
 
 | | Crystal Structure of B. subtilis ferrochelatase with Cobalt bound at the active site | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Ferrochelatase, ... | | Authors: | Soderberg, C.A.G, Hansson, M.D, Sreekanth, R, Al-Karadaghi, S, Hansson, M. | | Deposit date: | 2010-03-12 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Bacterial ferrochelatase goes human: Tyr13 determines the apparent metal specificity of Bacillus subtilis ferrochelatase
To be Published
|
|
8BLY
 
 | |
8B1K
 
 | | DtpB-Nb132-NV | | Descriptor: | ASN-VAL, DECANE, DODECANE, ... | | Authors: | Killer, M, Finocchio, G, Lei, J, Jungnickel, K, Kotov, V, Steinke, J, Bartels, K, Strauss, J, Dupeux, F, Humm, A.S, Cornaciu, I, Marquez, J, Pardon, E, Steyeart, J, Loew, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Plasticity of the binding pocket in peptide transporters underpins promiscuous substrate recognition.
Cell Rep, 42, 2023
|
|
8B1B
 
 | | DtpB-Nb132-AL | | Descriptor: | ALA-LEU, DECANE, DODECANE, ... | | Authors: | Killer, M, Finocchio, G, Lei, J, Jungnickel, K, Kotov, V, Steinke, J, Bartels, K, Strauss, J, Dupeux, F, Humm, A.S, Cornaciu, I, Marquez, J, Pardon, E, Steyeart, J, Loew, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Plasticity of the binding pocket in peptide transporters underpins promiscuous substrate recognition.
Cell Rep, 42, 2023
|
|
