1KO7
 
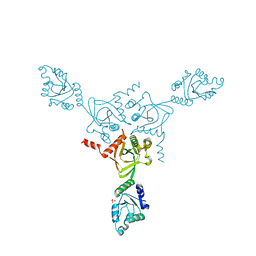 | | X-ray structure of the HPr kinase/phosphatase from Staphylococcus xylosus at 1.95 A resolution | | Descriptor: | Hpr kinase/phosphatase, PHOSPHATE ION | | Authors: | Marquez, J.A, Hasenbein, S, Koch, B, Fieulaine, S, Nessler, S, Hengstenberg, W, Scheffzek, K. | | Deposit date: | 2001-12-20 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the full-length HPr kinase/phosphatase from Staphylococcus xylosus at 1.95 A resolution: Mimicking the product/substrate of the phospho transfer reactions.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2HRO
 
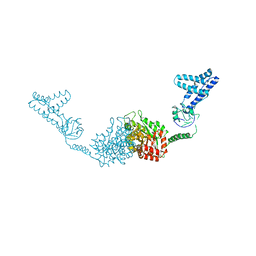 | | Structure of the full-lenght Enzyme I of the PTS system from Staphylococcus carnosus | | Descriptor: | Phosphoenolpyruvate-protein phosphotransferase, SULFATE ION | | Authors: | Marquez, J.A, Reinelt, S, Koch, B, Engelman, R, Hengstenberg, W, Scheffzek, K. | | Deposit date: | 2006-07-20 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the full-length enzyme I of the phosphoenolpyruvate-dependent sugar phosphotransferase system
J.Biol.Chem., 281, 2006
|
|
3QN1
 
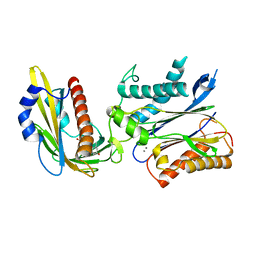 | | Crystal structure of the PYR1 Abscisic Acid receptor in complex with the HAB1 type 2C phosphatase catalytic domain | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1, MANGANESE (II) ION, ... | | Authors: | Betz, K, Dupeux, F, Santiago, J, Marquez, J.A. | | Deposit date: | 2011-02-07 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of Abscisic Acid Signaling in Vivo by an Engineered Receptor-Insensitive Protein Phosphatase Type 2C Allele.
Plant Physiol., 156, 2011
|
|
2NMS
 
 | |
3K90
 
 | | The Abscisic acid receptor PYR1 in complex with Abscisic Acid | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ACETIC ACID, GLYCEROL, ... | | Authors: | Dupeux, F.D, Santiago, J, Rodriguez, P.L, Marquez, J.A. | | Deposit date: | 2009-10-15 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The abscisic acid receptor PYR1 in complex with abscisic acid.
Nature, 462, 2009
|
|
7ZV8
 
 | | Crystal structure of SARS Cov-2 main protease in complex with an inhibitor 58 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, OCTANOIC ACID (CAPRYLIC ACID), ... | | Authors: | Rahimova, R, Di Micco, S, Marquez, J.A. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Rational design of the zonulin inhibitor AT1001 derivatives as potential anti SARS-CoV-2.
Eur.J.Med.Chem., 244, 2022
|
|
5A52
 
 | |
7ZV5
 
 | |
7ZV7
 
 | |
2X89
 
 | | Structure of the Beta2_microglobulin involved in amyloidogenesis | | Descriptor: | ANTIBODY, BETA-2-MICROGLOBULIN | | Authors: | Domanska, K, Srinivasan, V, Vanderhaegen, S, Pardon, E, Marquez, J.A, Bellotti, V, Wyns, L, Steyaert, J. | | Deposit date: | 2010-03-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Atomic Structure of a Nanobody-Trapped Domain-Swapped Dimer of an Amyloidogenic {Beta}2-Microglobulin Variant.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4QGK
 
 | |
2Q87
 
 | |
6YXG
 
 | | Cryogenic human adiponectin receptor 2 (ADIPOR2) with Tb-XO4 ligand determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, OLEIC ACID, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YXF
 
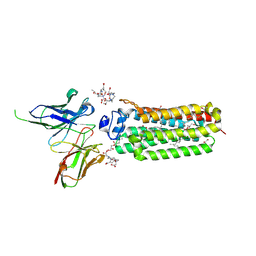 | | Cryogenic human adiponectin receptor 2 (ADIPOR2) with Gd-DO3 ligand determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, Adiponectin receptor protein 2, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.025 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YX9
 
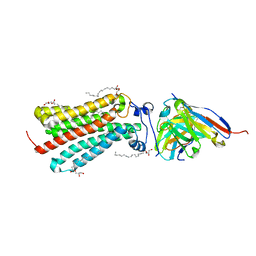 | | Cryogenic human adiponectin receptor 2 (ADIPOR2) at 2.4 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, GLYCEROL, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YXH
 
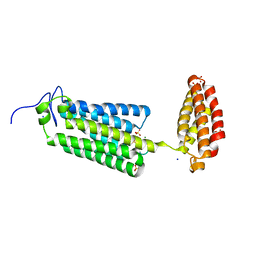 | | Cryogenic human alkaline ceramidase 3 (ACER3) at 2.6 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | Alkaline ceramidase 3, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YXD
 
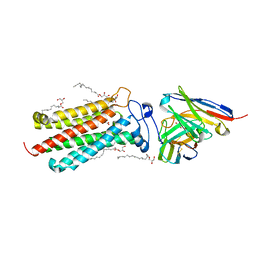 | | Room temperature structure of human adiponectin receptor 2 (ADIPOR2) at 2.9 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, GLYCEROL, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6H3R
 
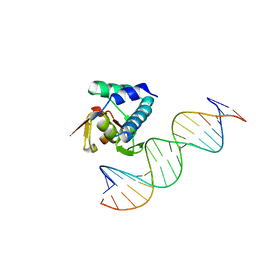 | |
6FZS
 
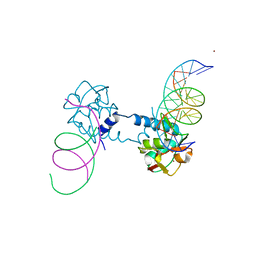 | | Crystal structure of Smad5-MH1 bound to the GGCGC site. | | Descriptor: | DNA (5'-D(P*TP*GP*CP*AP*GP*GP*CP*GP*CP*GP*CP*CP*TP*GP*CP*A)-3'), Mothers against decapentaplegic homolog 5, ZINC ION | | Authors: | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | Deposit date: | 2018-03-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Unveiling the dimer/monomer propensities of Smad MH1-DNA complexes
Computational and Structural Biotechnology Journal, 19, 2021
|
|
8B17
 
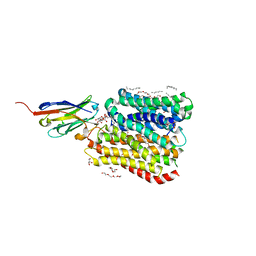 | | DtpB-Nb132-AWA | | Descriptor: | ALA-TRP-ALA, DECANE, DODECANE, ... | | Authors: | Killer, M, Finocchio, G, Lei, J, Jungnickel, K, Kotov, V, Steinke, J, Bartels, K, Strauss, J, Dupeux, F, Humm, A.S, Cornaciu, I, Marquez, J, Pardon, E, Steyeart, J, Loew, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Plasticity of the binding pocket in peptide transporters underpins promiscuous substrate recognition.
Cell Rep, 42, 2023
|
|
8B1D
 
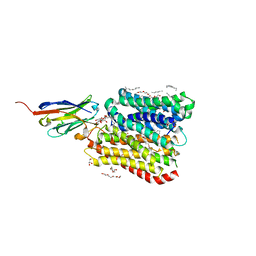 | | DtpB-Nb132-APF | | Descriptor: | ALA-PRO-PHE, DECANE, DODECANE, ... | | Authors: | Killer, M, Finocchio, G, Lei, J, Jungnickel, K, Kotov, V, Steinke, J, Bartels, K, Strauss, J, Dupeux, F, Humm, A.S, Cornaciu, I, Marquez, J, Pardon, E, Steyeart, J, Loew, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Plasticity of the binding pocket in peptide transporters underpins promiscuous substrate recognition.
Cell Rep, 42, 2023
|
|
8B1B
 
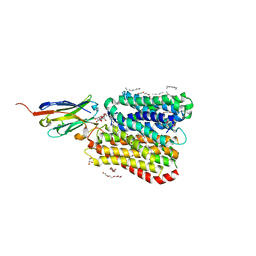 | | DtpB-Nb132-AL | | Descriptor: | ALA-LEU, DECANE, DODECANE, ... | | Authors: | Killer, M, Finocchio, G, Lei, J, Jungnickel, K, Kotov, V, Steinke, J, Bartels, K, Strauss, J, Dupeux, F, Humm, A.S, Cornaciu, I, Marquez, J, Pardon, E, Steyeart, J, Loew, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Plasticity of the binding pocket in peptide transporters underpins promiscuous substrate recognition.
Cell Rep, 42, 2023
|
|
8B19
 
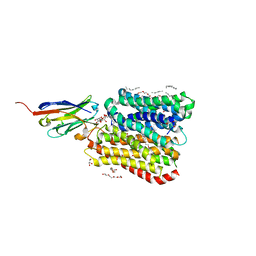 | | DtpB-Nb132-AFA | | Descriptor: | ALA-PHE-ALA, DECANE, DODECANE, ... | | Authors: | Killer, M, Finocchio, G, Lei, J, Jungnickel, K, Kotov, V, Steinke, J, Bartels, K, Strauss, J, Dupeux, F, Humm, A.S, Cornaciu, I, Marquez, J, Pardon, E, Steyeart, J, Loew, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Plasticity of the binding pocket in peptide transporters underpins promiscuous substrate recognition.
Cell Rep, 42, 2023
|
|
8B1I
 
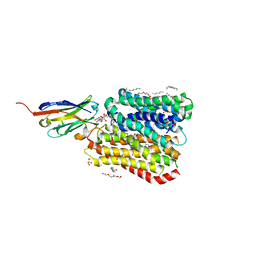 | | DtpB-Nb132-MS | | Descriptor: | DECANE, DODECANE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Killer, M, Finocchio, G, Lei, J, Jungnickel, K, Kotov, V, Steinke, J, Bartels, K, Strauss, J, Dupeux, F, Humm, A.S, Cornaciu, I, Marquez, J, Pardon, E, Steyeart, J, Loew, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Plasticity of the binding pocket in peptide transporters underpins promiscuous substrate recognition.
Cell Rep, 42, 2023
|
|
8B1K
 
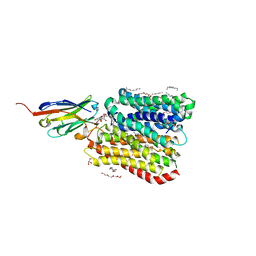 | | DtpB-Nb132-NV | | Descriptor: | ASN-VAL, DECANE, DODECANE, ... | | Authors: | Killer, M, Finocchio, G, Lei, J, Jungnickel, K, Kotov, V, Steinke, J, Bartels, K, Strauss, J, Dupeux, F, Humm, A.S, Cornaciu, I, Marquez, J, Pardon, E, Steyeart, J, Loew, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Plasticity of the binding pocket in peptide transporters underpins promiscuous substrate recognition.
Cell Rep, 42, 2023
|
|
