7OLH
 
 | |
1DBG
 
 | | CRYSTAL STRUCTURE OF CHONDROITINASE B | | Descriptor: | 4-deoxy-alpha-D-glucopyranose-(1-3)-[beta-D-glucopyranose-(1-4)]2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, CHONDROITINASE B | | Authors: | Huang, W, Matte, A, Li, Y, Kim, Y.S, Linhardt, R.J, Su, H, Cygler, M. | | Deposit date: | 1999-11-02 | | Release date: | 2000-01-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution.
J.Mol.Biol., 294, 1999
|
|
7OJR
 
 | |
1DEE
 
 | | Structure of S. aureus protein A bound to a human IgM Fab | | Descriptor: | IGM RF 2A2, IMMUNOGLOBULIN G BINDING PROTEIN A | | Authors: | Graille, M, Stura, E.A, Corper, A.L, Sutton, B.J, Taussig, M.J, Charbonnier, J.B, Silverman, G.J. | | Deposit date: | 1999-11-15 | | Release date: | 2000-05-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: structural basis for recognition of B-cell receptors and superantigen activity.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DS3
 
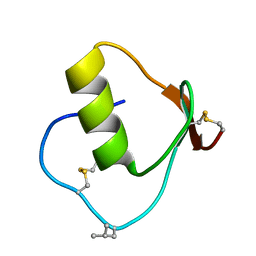 | | CRYSTAL STRUCTURE OF OMTKY3-CH2-ASP19I | | Descriptor: | OVOMUCOID | | Authors: | Bateman, K.S, Huang, K, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2000-01-06 | | Release date: | 2001-01-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Contribution of peptide bonds to inhibitor-protease binding: crystal structures of the turkey ovomucoid third domain backbone variants OMTKY3-Pro18I and OMTKY3-psi[COO]-Leu18I in complex with Streptomyces griseus proteinase B (SGPB) and the structure of the free inhibitor, OMTKY-3-psi[CH2NH2+]-Asp19I
J.Mol.Biol., 305, 2001
|
|
7OJS
 
 | |
7ORV
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00239 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORR
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00022 | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
1DF5
 
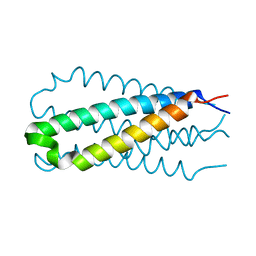 | |
3CPT
 
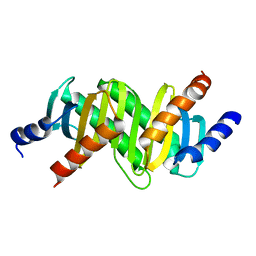 | | MP1-p14 Scaffolding complex | | Descriptor: | Mitogen-activated protein kinase kinase 1-interacting protein 1, Mitogen-activated protein-binding protein-interacting protein | | Authors: | Schrag, J.D, Cygler, M, Munger, C, Magloire, A. | | Deposit date: | 2008-04-01 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular dynamics-solvated interaction energy studies of protein-protein interactions: the MP1-p14 scaffolding complex.
J.Mol.Biol., 379, 2008
|
|
8V5Y
 
 | |
1E28
 
 | | Nonstandard peptide binding of HLA-B*5101 complexed with HIV immunodominant epitope KM2(TAFTIPSI) | | Descriptor: | BETA-2 MICROGLOBULIN LIGHT CHAIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN HEAVY CHAIN, PEPTIDE | | Authors: | Maenaka, K, Maenaka, T, Tomiyama, H, Takiguchi, M, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2000-05-18 | | Release date: | 2000-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Nonstandard peptide binding revealed by crystal structures of HLA-B*5101 complexed with HIV immunodominant epitopes.
J Immunol., 165, 2000
|
|
7TWC
 
 | | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to CAPS | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, DUF1479 domain-containing protein, GLYCEROL, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-02-07 | | Release date: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to CAPS
To Be Published
|
|
2OSQ
 
 | | NMR Structure of RRM-1 of Yeast NPL3 Protein | | Descriptor: | Nucleolar protein 3 | | Authors: | Deka, P, Bucheli, M, Skrisovska, L, Allain, F.H, Moore, C, Buratowski, S, Varani, G. | | Deposit date: | 2007-02-06 | | Release date: | 2007-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the yeast SR protein Npl3 and Interaction with mRNA 3'-end processing signals.
J.Mol.Biol., 375, 2008
|
|
7OUC
 
 | |
3OYS
 
 | | Human Carbonic Anhydrase II complexed with 2-{[4-AMINO-3-(3-HYDROXYPROP-1-YN-1-YL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-1-YL]METHYL}-5-METHYL-3-(2-METHYLPHENYL)QUINAZOLIN-4(3H)-ONE | | Descriptor: | 2-phenyl-N-(4-sulfamoylphenyl)acetamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Aggarwal, M, McKenna, R. | | Deposit date: | 2010-09-23 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.538 Å) | | Cite: | Anticonvulsant 4-aminobenzenesulfonamide derivatives with branched-alkylamide moieties: X-ray crystallography and inhibition studies of human carbonic anhydrase isoforms I, II, VII, and XIV.
J.Med.Chem., 54, 2011
|
|
7TB3
 
 | | cryo-EM structure of MBP-KIX-apoferritin | | Descriptor: | Isoform 2 of CREB-binding protein,Ferritin heavy chain, N-terminally processed | | Authors: | Zhang, K, Horikoshi, N, Li, S, Powers, A, Hameedi, M, Pintilie, G, Chae, H, Khan, Y, Suomivuori, C, Dror, R, Sakamoto, K, Chiu, W, Wakatsuki, S. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Cryo-EM, Protein Engineering, and Simulation Enable the Development of Peptide Therapeutics against Acute Myeloid Leukemia.
Acs Cent.Sci., 8, 2022
|
|
7OUD
 
 | |
7OUJ
 
 | | Crystal structure of the flavoprotein monooxygenase RubL from rubromycin biosynthesis | | Descriptor: | (2S)-hexane-1,2,6-triol, 4-HYDROXYPROLINE, CHLORIDE ION, ... | | Authors: | Saleem-Batcha, R, Toplak, M, Teufel, R. | | Deposit date: | 2021-06-11 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Catalytic Control of Spiroketal Formation in Rubromycin Polyketide Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7P06
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in outward-facing conformation with ADP-orthovanadate/ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADP ORTHOVANADATE, MAGNESIUM ION, ... | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P05
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP and rhodamine 6G | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Pleiotropic ABC efflux transporter of multiple drugs, ... | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
1DAU
 
 | | Analog of dickerson-drew DNA dodecamer with 6'-alpha-methyl carbocyclic thymidines, NMR, minimized average structure | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*(T32)P*(T32)P*CP*GP*CP*G)-3') | | Authors: | Denisov, A, Bekiroglu, S, Maltseva, T, Sandstrom, A, Altmann, K.-H, Egli, M, Chattopadhyaya, J. | | Deposit date: | 1998-01-21 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution conformation of a carbocyclic analog of the Dickerson-Drew dodecamer: comparison with its own X-ray structure and that of the NMR structure of the native counterpart.
J.Biomol.Struct.Dyn., 16, 1998
|
|
7TBH
 
 | | cryo-EM structure of MBP-KIX-apoferritin complex with peptide 7 | | Descriptor: | Isoform 2 of CREB-binding protein,Ferritin heavy chain, N-terminally processed, LEU-SER-ARG-ARG-PRO-SEP-TYR-ARG-LYS-ILE-LEU-ASN-ASP-LEU-SER-SER-ASP-ALA-PRO | | Authors: | Zhang, K, Horikoshi, N, Li, S, Powers, A, Hameedi, M, Pintilie, G, Chae, H, Khan, Y, Suomivuori, C, Dror, R, Sakamoto, K, Chiu, W, Wakatsuki, S. | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM, Protein Engineering, and Simulation Enable the Development of Peptide Therapeutics against Acute Myeloid Leukemia.
Acs Cent.Sci., 8, 2022
|
|
1DM1
 
 | | 2.0 A CRYSTAL STRUCTURE OF THE DOUBLE MUTANT H(E7)V, T(E10)R OF MYOGLOBIN FROM APLYSIA LIMACINA | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Federici, L, Savino, C, Musto, R, Travaglini-Allocatelli, C, Cutruzzola, F, Brunori, M. | | Deposit date: | 1999-12-13 | | Release date: | 2000-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering His(E7) affects the control of heme reactivity in Aplysia limacina myoglobin.
Biochem.Biophys.Res.Commun., 269, 2000
|
|
6FK1
 
 | | Cyclophilin A | | Descriptor: | 1,2-ETHANEDIOL, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Lisboa, J, dos Santos, N.M.S. | | Deposit date: | 2018-01-23 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Production, crystallization and structure determination of cyclophilin A from Mus musculus
To Be Published
|
|
