2WNY
 
 | | Structure of Mth689, a DUF54 protein from Methanothermobacter thermautotrophicus | | Descriptor: | CHLORIDE ION, CONSERVED PROTEIN MTH689, NICKEL (II) ION | | Authors: | Ng, C.L, Waterman, D.G, Lebedev, A.A, Smits, C, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2009-07-21 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Mth689, a Duf54 Protein from Methanothermobacter Thermautotrophicus
To be Published
|
|
1XFA
 
 | | Structure of NBD1 from murine CFTR- F508R mutant | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Thibodeau, P.H, Brautigam, C.A, Machius, M, Thomas, P.J. | | Deposit date: | 2004-09-14 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Side chain and backbone contributions of Phe508 to CFTR folding.
Nat.Struct.Mol.Biol., 12, 2005
|
|
3OS9
 
 | | Estrogen Receptor | | Descriptor: | 4-[1-allyl-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor | | Authors: | Bruning, J, Parent, A.A, Gil, G, Zhao, M, Nowak, J, Pace, M.C, Smith, C.L, Afonine, P.V, Adams, P.D, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2010-09-08 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Coupling of receptor conformation and ligand orientation determine graded activity.
Nat.Chem.Biol., 6, 2010
|
|
1XI9
 
 | | Alanine aminotransferase from Pyrococcus furiosus Pfu-1397077-001 | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, putative transaminase | | Authors: | Zhou, W, Tempel, W, Shah, A, Chen, L, Liu, Z.-J, Lee, D, Lin, D, Chang, S.-H, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Lee, H.-S, Poole II, F.L, Shah, C, Sugar, F.J, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Alanine aminotransferase from Pyrococcus furiosus Pfu-1397077-001
To be published
|
|
3HNG
 
 | | Crystal structure of VEGFR1 in complex with N-(4-Chlorophenyl)-2-((pyridin-4-ylmethyl)amino)benzamide | | Descriptor: | CHLORIDE ION, N-(4-chlorophenyl)-2-[(pyridin-4-ylmethyl)amino]benzamide, Vascular endothelial growth factor receptor 1 | | Authors: | Tresaugues, L, Roos, A, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Kragh-Nielsen, T, Kotzch, A, Sagemark, J, Schueler, H, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Van der Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-31 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of VEGFR1 in complex with N-(4-Chlorophenyl)-2-((pyridin-4-ylmethyl)amino)benzamide
To be Published
|
|
1XJC
 
 | | X-ray crystal structure of MobB protein homolog from Bacillus stearothermophilus | | Descriptor: | MobB protein homolog | | Authors: | Osipiuk, J, Zhou, M, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-23 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of MobB protein homolog from Bacillus stearothermophilus
To be Published
|
|
1XH3
 
 | | Conformational Restraints and Flexibility of 14-Meric Peptides in Complex with HLA-B*3501 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Probst-Kepper, M, Hecht, H.J, Herrmann, H, Janke, V, Ocklenburg, F, Klempnauer, J, van den Eynde, B.J, Weiss, S. | | Deposit date: | 2004-09-17 | | Release date: | 2004-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Conformational restraints and flexibility of 14-meric peptides in complex with HLA-B*3501.
J.Immunol., 173, 2004
|
|
4H4S
 
 | | Crystal Structure of Ferredoxin reductase, BphA4 E175C/Q177G mutant (reduced form) | | Descriptor: | Biphenyl dioxygenase ferredoxin reductase subunit, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Nishizawa, A, Harada, A, Senda, M, Tachihara, Y, Muramatsu, D, Kishigami, S, Mori, S, Sugiyama, K, Senda, T, Kimura, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Random Mutagenesis with the Project Assessment for Complete Conversion of Co-Factor Specificity of a Ferredoxin Reductase BphA4
To be Published
|
|
2VR0
 
 | |
4H0Z
 
 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with N-acetyl muramic acid at 2.0 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-beta-muramic acid, rRNA N-glycosidase | | Authors: | Singh, A, Pandey, S, Kushwaha, G.S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-09-10 | | Release date: | 2012-10-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with N-acetyl muramic acid at 2.0 Angstrom resolution
TO BE PUBLISHED
|
|
2VYR
 
 | | Structure of human MDM4 N-terminal domain bound to a single domain antibody | | Descriptor: | HUMAN SINGLE DOMAIN ANTIBODY, MDM4 PROTEIN, SULFATE ION | | Authors: | Yu, G.W, Vaysburd, M, Allen, M.D, Settanni, G, Fersht, A.R. | | Deposit date: | 2008-07-28 | | Release date: | 2008-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Human Mdm4 N-Terminal Domain Bound to a Single-Domain Antibody.
J.Mol.Biol., 385, 2009
|
|
4H2F
 
 | | Human ecto-5'-nucleotidase (CD73): crystal form I (open) in complex with adenosine | | Descriptor: | 5'-nucleotidase, ADENOSINE, CALCIUM ION, ... | | Authors: | Straeter, N, Knapp, K.M, Zebisch, M, Pippel, J. | | Deposit date: | 2012-09-12 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Human Ecto-5'-Nucleotidase (CD73): Insights into the Regulation of Purinergic Signaling.
Structure, 20, 2012
|
|
2VXA
 
 | | H. halophila dodecin in complex with riboflavin | | Descriptor: | CHLORIDE ION, DODECIN, RIBOFLAVIN | | Authors: | Grininger, M, Staudt, H, Johansson, P, Wachtveitl, J, Oesterhelt, D. | | Deposit date: | 2008-07-01 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dodecin is the Key Player in Flavin Homeostasis of Archaea.
J.Biol.Chem., 284, 2009
|
|
3OXZ
 
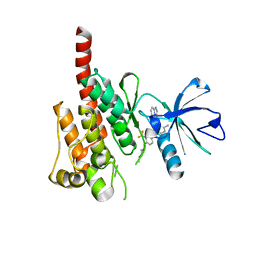 | | Crystal structure of ABL kinase domain bound with a DFG-out inhibitor AP24534 | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Tyrosine-protein kinase ABL1 | | Authors: | Zhou, T, Huang, W.S, Wang, Y, Thomas, M, Keats, J, Xu, Q, Rivera, V, Shakespeare, W.C, Clackson, T, Dalgarno, D.C, Zhu, X. | | Deposit date: | 2010-09-22 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib (AP24534): Lessons for Overcoming Kinase Inhibitor Resistance.
Chem.Biol.Drug Des., 77, 2011
|
|
4H63
 
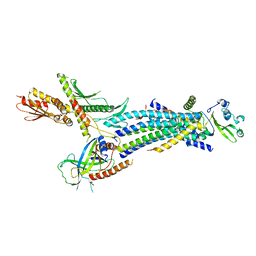 | | Structure of the Schizosaccharomyces pombe Mediator head module | | Descriptor: | Mediator of RNA polymerase II transcription subunit 11, Mediator of RNA polymerase II transcription subunit 17, Mediator of RNA polymerase II transcription subunit 18, ... | | Authors: | Lariviere, L, Plaschka, C, Seizl, M, Wenzeck, L, Kurth, F, Cramer, P. | | Deposit date: | 2012-09-19 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of the Mediator head module.
Nature, 492, 2012
|
|
4H33
 
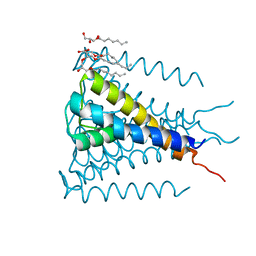 | | Crystal structure of a voltage-gated K+ channel pore module in a closed state in lipid membranes, tetragonal crystal form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lmo2059 protein, POTASSIUM ION | | Authors: | Santos, J.S, Asmar-Rovira, G.A, Han, G.W, Liu, W, Syeda, R, Cherezov, V, Baker, K.A, Stevens, R.C, Montal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Voltage-gated K+ Channel Pore Module in a Closed State in Lipid Membranes.
J.Biol.Chem., 287, 2012
|
|
8CDA
 
 | | Crystal structure of MAB_4123 from Mycobacterium abscessus | | Descriptor: | GLYCEROL, PHOSPHATE ION, Probable monooxygenase | | Authors: | Ung, K.L, Poussineau, C, Couston, J, Alsarraf, H.M.A.B, Blaise, M. | | Deposit date: | 2023-01-30 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of MAB_4123, a putative flavin-dependent monooxygenase from Mycobacterium abscessus.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8BV2
 
 | |
4AKD
 
 | | High resolution structure of Mannose Binding lectin from Champedak (CMB) | | Descriptor: | CADMIUM ION, CHLORIDE ION, MANNOSE-SPECIFIC LECTIN KM+ | | Authors: | Gabrielsen, M, Abdul-Rahman, P.S, Othman, S, Hashim, O.H, Cogdell, R.J. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures and Binding Specificity of Galactose- and Mannose-Binding Lectins from Champedak: Differences from Jackfruit Lectins
Acta Crystallogr.,Sect.F, 70, 2014
|
|
5B7M
 
 | | Structure of perdeuterated CueO - the signal peptide was truncated by HRV3C protease | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION | | Authors: | Akter, M, Higuchi, Y, Shibata, N. | | Deposit date: | 2016-06-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical, spectroscopic and X-ray structural analysis of deuterated multicopper oxidase CueO prepared from a new expression construct for neutron crystallography
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1XP0
 
 | | Catalytic Domain Of Human Phosphodiesterase 5A In Complex With Vardenafil | | Descriptor: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-07 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
4AK4
 
 | | High resolution structure of Galactose Binding lectin from Champedak (CGB) | | Descriptor: | AGGLUTININ ALPHA CHAIN, AGGLUTININ BETA-4 CHAIN, HEXAETHYLENE GLYCOL | | Authors: | Gabrielsen, M, Abdul-Rahman, P.S, Othman, S, Hashim, O.H, Cogdell, R.J. | | Deposit date: | 2012-02-21 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures and Binding Specificity of Galactose- and Mannose-Binding Lectins from Champedak: Differences from Jackfruit Lectins
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4H2G
 
 | | Human ecto-5'-nucleotidase (CD73): crystal form II (open) in complex with adenosine | | Descriptor: | 5'-nucleotidase, ADENOSINE, CALCIUM ION, ... | | Authors: | Straeter, N, Knapp, K.M, Zebisch, M, Pippel, J. | | Deposit date: | 2012-09-12 | | Release date: | 2012-11-28 | | Last modified: | 2012-12-26 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the Human Ecto-5'-Nucleotidase (CD73): Insights into the Regulation of Purinergic Signaling.
Structure, 20, 2012
|
|
1XJX
 
 | | The crystal structures of the DNA binding sites of the RUNX1 transcription factor | | Descriptor: | 5'-D(*TP*CP*TP*GP*CP*GP*GP*TP*C)-3', 5'-D(*TP*GP*AP*CP*CP*GP*CP*AP*G)-3' | | Authors: | Kitayner, M, Rozenberg, H, Rabinovich, D, Shakked, Z. | | Deposit date: | 2004-09-26 | | Release date: | 2005-03-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the DNA-binding site of Runt-domain transcription regulators.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2WAG
 
 | | The Structure of a family 25 Glycosyl hydrolase from Bacillus anthracis. | | Descriptor: | GLYCEROL, LYSOZYME, PUTATIVE, ... | | Authors: | Martinez-Fleites, C, Korczynska, J.E, Cope, M, Turkenburg, J.P, Taylor, E.J. | | Deposit date: | 2009-02-06 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of a Family Gh25 Lysozyme from Bacillus Anthracis Implies a Neighboring-Group Catalytic Mechanism with Retention of Anomeric Configuration
Carbohydr.Res., 344, 2009
|
|
