7KEP
 
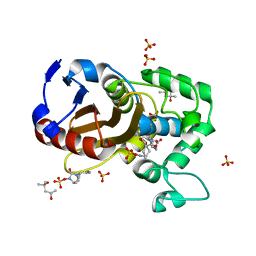 | | avibactam-CDD-1 2 minute complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (2S,5R)-7-oxo-6-(sulfooxy)-1,6-diazabicyclo[3.2.1]octane-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-10-11 | | Release date: | 2021-01-20 | | Last modified: | 2021-05-26 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Inhibition of the Clostridioides difficile Class D beta-Lactamase CDD-1 by Avibactam.
Acs Infect Dis., 7, 2021
|
|
1A3H
 
 | |
7KWG
 
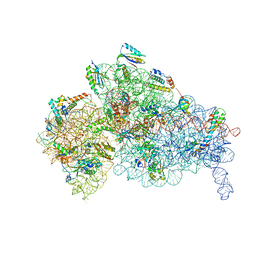 | | Staphylococcus aureus 30S ribosomal subunit in presence of spermidine | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Belinite, M, Khusainov, I, Marzi, S, Romby, P, Yusupov, M, Hashem, Y. | | Deposit date: | 2020-11-30 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Stabilization of Ribosomal RNA of the Small Subunit by Spermidine in Staphylococcus aureus .
Front Mol Biosci, 8, 2021
|
|
4K8F
 
 | |
1J0Z
 
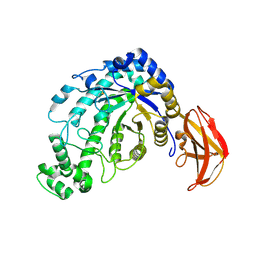 | | Beta-amylase from Bacillus cereus var. mycoides in complex with maltose | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1J0Y
 
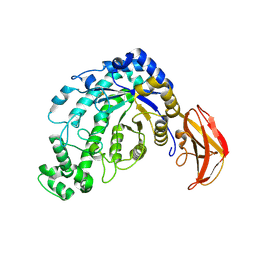 | | Beta-amylase from Bacillus cereus var. mycoides in complex with glucose | | Descriptor: | Beta-amylase, CALCIUM ION, beta-D-glucopyranose | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
6VJ2
 
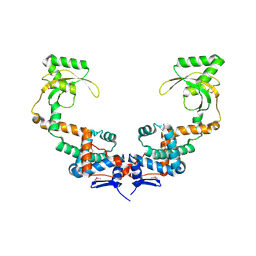 | | 3.10 Angstrom Resolution Crystal Structure of Foldase Protein (PrsA) from Lactococcus lactis | | Descriptor: | Foldase | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-14 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | 3.10 Angstrom Resolution Crystal Structure of Foldase Protein (PrsA) from Lactococcus lactis
To Be Published
|
|
4K9Y
 
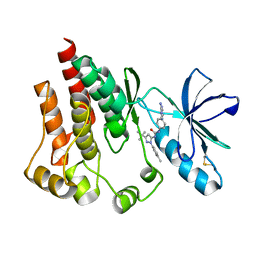 | | FOCAL ADHESION KINASE Catalytic domain in complex with 1-[4-(6-Amino-purin-9-yl)-phenyl]-3-(5-tert-butyl-2-p-tolyl-2H-pyrazol-3-yl)-urea | | Descriptor: | 1-[4-(6-amino-9H-purin-9-yl)phenyl]-3-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]urea, Focal adhesion kinase 1 | | Authors: | Musil, D, Graedler, U, Lehmann, M, Heinrich, T, Dresing, V. | | Deposit date: | 2013-04-21 | | Release date: | 2013-09-11 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based discovery of focal adhesion kinase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6VZ3
 
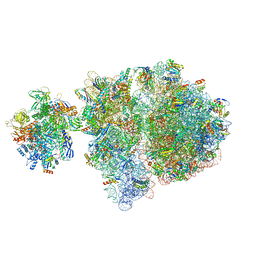 | | Escherichia coli transcription-translation complex D2 (TTC-D2) containing mRNA with a 27 nt long spacer | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
1J2F
 
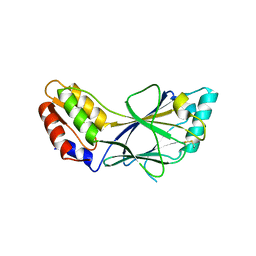 | | X-ray crystal structure of IRF-3 and its functional implications | | Descriptor: | Interferon regulatory factor 3 | | Authors: | Takahasi, K, Noda, N, Horiuchi, M, Mori, M, Okabe, Y, Fukuhara, Y, Terasawa, H, Fujita, T, Inagaki, F. | | Deposit date: | 2003-01-04 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of IRF-3 and its functional implications.
Nat.Struct.Biol., 10, 2003
|
|
7L0Q
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6VKS
 
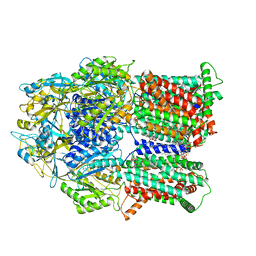 | | Cryo-electron microscopy structures of a gonococcal multidrug efflux pump illuminate a mechanism of drug recognition with ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Efflux pump membrane transporter, PHOSPHATIDYLETHANOLAMINE | | Authors: | Moseng, M.A, Lyu, M. | | Deposit date: | 2020-01-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM Structures of a Gonococcal Multidrug Efflux Pump Illuminate a Mechanism of Drug Recognition and Resistance.
Mbio, 11, 2020
|
|
1J2W
 
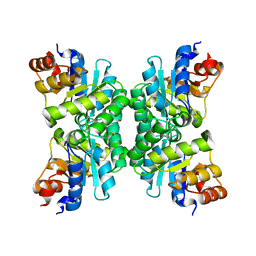 | | Tetrameric Structure of aldolase from Thermus thermophilus HB8 | | Descriptor: | Aldolase protein | | Authors: | Lokanath, N.K, Shiromizu, I, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-14 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6VS2
 
 | | protein D | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, Multidrug transporter MdfA, PRASEODYMIUM ION | | Authors: | Lu, M, Lu, M.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and mechanism of a redesigned multidrug transporter from the Major Facilitator Superfamily.
Sci Rep, 10, 2020
|
|
6W17
 
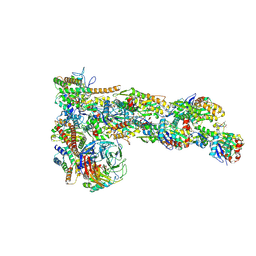 | | Structure of Dip1-activated Arp2/3 complex with nucleated actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Shaaban, M, Nolen, B.J, Chowdhury, S. | | Deposit date: | 2020-03-03 | | Release date: | 2020-08-12 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM reveals the transition of Arp2/3 complex from inactive to nucleation-competent state.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VVZ
 
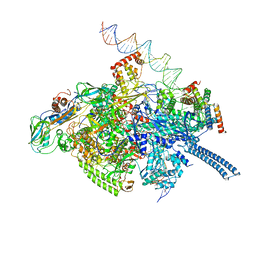 | | Mycobacterium tuberculosis RNAP S456L mutant transcription initiation intermediate structure with Sorangicin | | Descriptor: | DNA (62-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2JY8
 
 | |
8PIE
 
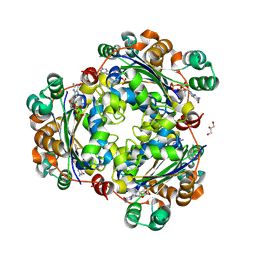 | | Crystal structure of the human nucleoside diphosphate kinase B domain in complex with the product AT-8500 formed by catalysis of compound AT-9010 | | Descriptor: | GLYCEROL, Nucleoside diphosphate kinase B, [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-fluoranyl-4-methyl-3-oxidanyl-oxolan-2-yl]methyl phosphono hydrogen phosphate | | Authors: | Feracci, M, Chazot, A, Ferron, F, Alvarez, K, Canard, B. | | Deposit date: | 2023-06-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The activation cascade of the broad-spectrum antiviral bemnifosbuvir characterized at atomic resolution.
Plos Biol., 22, 2024
|
|
3QR8
 
 | |
5MGI
 
 | | Crystal structure of KPC-2 carbapenemase in complex with a phenyl boronic inhibitor. | | Descriptor: | (~{E})-3-[2-(dihydroxyboranyl)phenyl]prop-2-enoic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Vicario, M, Celenza, G, Bellio, P, Perilli, M.G, Tondi, D, Cendron, L. | | Deposit date: | 2016-11-21 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phenylboronic Acid Derivatives as Validated Leads Active in Clinical Strains Overexpressing KPC-2: A Step against Bacterial Resistance.
Chemmedchem, 13, 2018
|
|
2K1G
 
 | | Solution NMR structure of lipoprotein spr from Escherichia coli K12. Northeast Structural Genomics target ER541-37-162 | | Descriptor: | Lipoprotein spr | | Authors: | Aramini, J.M, Rossi, P, Zhao, L, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Huang, Y.J, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-03 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the NlpC/P60 domain of lipoprotein Spr from Escherichia coli: structural evidence for a novel cysteine peptidase catalytic triad.
Biochemistry, 47, 2008
|
|
5MKF
 
 | | cryoEM Structure of Polycystin-2 in complex with calcium and lipids | | Descriptor: | 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wilkes, M, Madej, M.G, Ziegler, C. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular insights into lipid-assisted Ca(2+) regulation of the TRP channel Polycystin-2.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4PGO
 
 | | Crystal structure of hypothetical protein PF0907 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | Descriptor: | CHLORIDE ION, Uncharacterized protein | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-05-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
6VP0
 
 | | Human Diacylglycerol Acyltransferase 1 in complex with oleoyl-CoA | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Diacylglycerol O-acyltransferase 1, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Wang, L, Qian, H, Han, Y, Nian, Y, Ren, Z, Zhang, H, Hu, L, Prasad, B.V.V, Yan, N, Zhou, M. | | Deposit date: | 2020-02-01 | | Release date: | 2020-05-13 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and mechanism of human diacylglycerol O-acyltransferase 1.
Nature, 581, 2020
|
|
4L37
 
 | | SP2-SP3 - a complex of two storage proteins from Bombyx mori hemolymph | | Descriptor: | Arylphorin, SODIUM ION, SULFATE ION, ... | | Authors: | Pietrzyk, A.J, Bujacz, A, Mueller-Dieckmann, J, Jaskolski, M, Bujacz, G. | | Deposit date: | 2013-06-05 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic identification of an unexpected protein complex in silkworm haemolymph.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
