8DVX
 
 | | Structure of acetylated Pig somatic Cytochrome c (Aly39) at 1.5A | | Descriptor: | Cytochrome c, HEME C, HEXACYANOFERRATE(3-) | | Authors: | Edwards, B.F.P, Huettemann, M, Vaishnav, A, Brunzelle, J, Morse, P, Wan, J. | | Deposit date: | 2022-07-30 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cytochrome c lysine acetylation regulates cellular respiration and cell death in ischemic skeletal muscle.
Nat Commun, 14, 2023
|
|
2SGD
 
 | | ASP 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 10.7 | | Descriptor: | Ovomucoid, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pHs
To be Published
|
|
7ATS
 
 | | The LIMK1 Kinase Domain Bound To LIJTF500127 | | Descriptor: | LIM domain kinase 1, N-[3-[5-(4-Chlorophenyl)-1H-pyrrolo[2,3-b]pyridine-3-carbonyl]-2,4-difluorophenyl]benzenesulfonamide | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Yamamoto, S, Tawada, M, Nomura, I, Takagi, T, Ahmed, M, Little, W, Mueller-Knapp, S, Knapp, S. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The LIMK1 Kinase Domain Bound To LIJTF500127
To Be Published
|
|
8DZL
 
 | | Structure of the K39Q mutant of rat somatic Cytochrome c at 1.36A | | Descriptor: | Cytochrome c, somatic, HEME C | | Authors: | Edwards, B.F.P, Huettemann, M, Vaishnav, A, Brunzelle, J, Morse, P, Wan, J. | | Deposit date: | 2022-08-08 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Cytochrome c lysine acetylation regulates cellular respiration and cell death in ischemic skeletal muscle.
Nat Commun, 14, 2023
|
|
6TTG
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with LMD62 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-(2-morpholin-4-ylethoxy)-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, ... | | Authors: | Welin, M, Kimbung, R, Focht, D. | | Deposit date: | 2019-12-27 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | New dual ATP-competitive inhibitors of bacterial DNA gyrase and topoisomerase IV active against ESKAPE pathogens.
Eur.J.Med.Chem., 213, 2021
|
|
6I0L
 
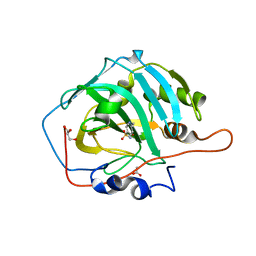 | | Crystal structure of human carbonic anhydrase I in complex with the 1-[4-chloro-3-(trifluoromethyl)phenyl]-3-[2-(4-sulfamoylphenyl)ethyl]urea inhibitor | | Descriptor: | 1-[4-chloranyl-3-(trifluoromethyl)phenyl]-3-[2-(4-sulfamoylphenyl)ethyl]urea, ACETATE ION, Carbonic anhydrase 1, ... | | Authors: | Ferraroni, M, Supuran, C.T, Bozdag, M, Chiapponi, D. | | Deposit date: | 2018-10-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Carbonic anhydrase inhibitors based on sorafenib scaffold: Design, synthesis, crystallographic investigation and effects on primary breast cancer cells.
Eur.J.Med.Chem., 182, 2019
|
|
2RS4
 
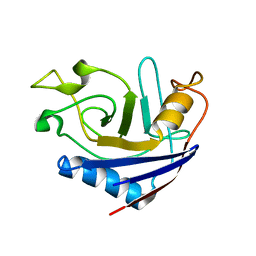 | | NMR structure of stereo-array isotope labelled (SAIL) peptidyl-prolyl cis-trans isomerase from E. coli (EPPIb) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase B | | Authors: | Takeda, M, Jee, J, Ono, A.M, Okuma, K, Terauchi, T, Kainosho, M. | | Deposit date: | 2011-07-16 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Hydrogen exchange study on the hydroxyl groups of serine and threonine residues in proteins and structure refinement using NOE restraints with polar side-chain groups
J.Am.Chem.Soc., 133, 2011
|
|
7B60
 
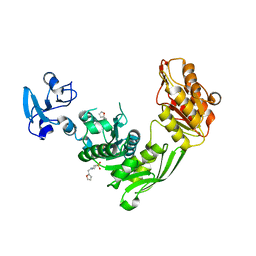 | | Crystal structure of MurE from E.coli in complex with Z1269139261 | | Descriptor: | 4-[(furan-2-yl)methyl]-1lambda~6~,4-thiazinane-1,1-dione, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
6JND
 
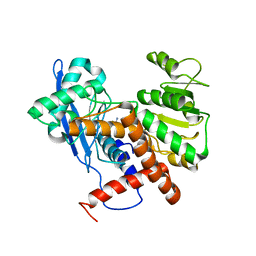 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | Authors: | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | Deposit date: | 2019-03-14 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
1X68
 
 | | Solution structures of the C-terminal LIM domain of human FHL5 protein | | Descriptor: | FHL5 protein, ZINC ION | | Authors: | Nameki, N, Sasagawa, A, Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the C-terminal LIM domain of human FHL5 protein
To be Published
|
|
6TMY
 
 | | Crystal structure of isoform CBd of the basic phospholipase A2 subunit of crotoxin from Crotalus durissus terrificus | | Descriptor: | CHLORIDE ION, Phospholipase A2 crotoxin basic subunit CBc, SODIUM ION, ... | | Authors: | Nemecz, D, Ostrowski, M, Saul, F.A, Faure, G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Isoform CBd of the Basic Phospholipase A 2 Subunit of Crotoxin: Description of the Structural Framework of CB for Interaction with Protein Targets.
Molecules, 25, 2020
|
|
7B6M
 
 | | Crystal structure of MurE from E.coli in complex with Z1198948504 | | Descriptor: | 1,2-ETHANEDIOL, N-[2-(2,5-Dioxopyrrolidin-1-yl)ethyl]-3-methylbenzamide, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC), Structural Genomics Consortium for Research on Gene Expression (SGCGES) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
2RQB
 
 | | Solution structure of MDA5 CTD | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, ZINC ION | | Authors: | Takahasi, K, Kumeta, H, Tsuduki, N, Narita, R, Shigemoto, T, Hirai, R, Yoneyama, M, Horiuchi, M, Ogura, K, Fujita, T, Fuyuhiko, I. | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-05 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Cytosolic RNA Sensor MDA5 and LGP2 C-terminal Domains: IDENTIFICATION OF THE RNA RECOGNITION LOOP IN RIG-I-LIKE RECEPTORS
J.Biol.Chem., 284, 2009
|
|
2RNG
 
 | | Solution structure of big defensin | | Descriptor: | Big defensin | | Authors: | Kouno, T, Fujitani, N, Osaki, T, Kawabata, S, Nishimura, S, Mizuguchi, M, Aizawa, T, Demura, M, Nitta, K, Kawano, K. | | Deposit date: | 2007-12-27 | | Release date: | 2008-10-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A novel beta-defensin structure: a potential strategy of big defensin for overcoming resistance by Gram-positive bacteria
Biochemistry, 47, 2008
|
|
2BPM
 
 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-630529 | | Descriptor: | (2S)-N-[(3Z)-5-CYCLOPROPYL-3H-PYRAZOL-3-YLIDENE]-2-[4-(2-OXOIMIDAZOLIDIN-1-YL)PHENYL]PROPANAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Cameron, A, Fogliatto, G, Pevarello, P, Brasca, M.G, Orsini, P, Traquandi, G, Longo, A, Nesi, M, Orzi, F, Piutti, C, Sansonna, P, Varasi, M, Vulpetti, A, Roletto, F, Alzani, R, Ciomei, M, Albanese, C, Pastori, W, Marsiglio, A, Pesenti, E, Fiorentini, F, Bischoff, J.R, Mercurio, C. | | Deposit date: | 2005-04-21 | | Release date: | 2005-12-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3-Aminopyrazole Inhibitors of Cdk2-Cyclin a as Antitumor Agents. 2. Lead Optimization
J.Med.Chem., 48, 2005
|
|
6FQ5
 
 | | Class 1 : canonical nucleosome | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B, ... | | Authors: | Bilokapic, S, Halic, M. | | Deposit date: | 2018-02-13 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural rearrangements of the histone octamer translocate DNA.
Nat Commun, 9, 2018
|
|
6G5A
 
 | | Heme-carbene complex in myoglobin H64V/V68A containing an N-methylhistidine as the proximal ligand, 1.48 angstrom resolution | | Descriptor: | ETHYL ACETATE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tinzl, M, Hayashi, T, Mori, T, Hilvert, D. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | Capture and characterization of a reactive haem-carbenoid complex in an artificial metalloenzyme
Nat Catal, 1, 2018
|
|
6FRW
 
 | | X-ray structure of the levansucrase from Erwinia tasmaniensis | | Descriptor: | GLYCEROL, Levansucrase (Beta-D-fructofuranosyl transferase), ZINC ION | | Authors: | Polsinelli, I, Salomone-Stagni, M, Caliandro, R, Demitri, N, Benini, S. | | Deposit date: | 2018-02-16 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Comparison of the Levansucrase from the epiphyte Erwinia tasmaniensis vs its homologue from the phytopathogen Erwinia amylovora.
Int. J. Biol. Macromol., 127, 2019
|
|
2RMJ
 
 | | Solution structure of RIG-I C-terminal domain | | Descriptor: | Probable ATP-dependent RNA helicase DDX58 | | Authors: | Takahasi, K, Yoneyama, M, Nihishori, T, Hirai, R, Narita, R, Gale Jr, M, Fujita, T, Inagaki, F. | | Deposit date: | 2007-10-23 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nonself RNA-Sensing Mechanism of RIG-I Helicase and Activation of Antiviral Immune Responses
Mol.Cell, 29, 2008
|
|
6JYP
 
 | | Crystal structure of uPA_H99Y in complex with 3-azanyl-5-(azepan-1-yl)-N-[bis(azanyl)methylidene]-6-chloranyl-pyrazine-2-carboxamide | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-N-[bis(azanyl)methylidene]-6-chloranyl-pyrazine-2-carboxamide, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Buckley, B, Jiang, L.G, Huang, M.D, Kelso, M, Ranson, M. | | Deposit date: | 2019-04-27 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | uPA-HMA
To Be Published
|
|
1X80
 
 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
7B6N
 
 | | Crystal structure of MurE from E.coli in complex with Z757284952 | | Descriptor: | 1,2-ETHANEDIOL, N-(2-(2,4-dioxothiazolidin-3-yl)ethyl)-3-methylbenzamide, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
8GM3
 
 | | Vibrio harveyi Holo HphA | | Descriptor: | HEME B/C, Hemophilin | | Authors: | Pan, C, Shah, M, Moraes, T.F. | | Deposit date: | 2023-03-24 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.727 Å) | | Cite: | Prevalence of Slam-dependent hemophilins in Gram-negative bacteria.
J.Bacteriol., 206, 2024
|
|
6K0H
 
 | | Crystal Structure of UDP-glucose 4-epimerase from Bifidobacterium longum in complex with NAD+ and UDP-GlcNAc | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Nam, Y.-W, Nishimoto, M, Arakawa, T, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2019-05-06 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for broad substrate specificity of UDP-glucose 4-epimerase in the human milk oligosaccharide catabolic pathway of Bifidobacterium longum.
Sci Rep, 9, 2019
|
|
5LTF
 
 | | Crystal structure of Lymphocytic choriomeningitis mammarenavirus endonuclease complexed with catalytic ions | | Descriptor: | MAGNESIUM ION, RNA-directed RNA polymerase L | | Authors: | Saez-Ayala, M, Yekwa, E.L, Canard, B, Ferron, F. | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structures ofLymphocytic choriomeningitis virusendonuclease domain complexed with diketo-acid ligands.
IUCrJ, 5, 2018
|
|
