5CPT
 
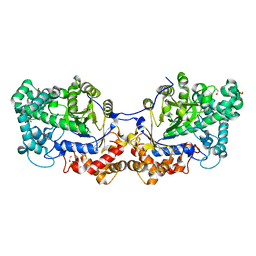 | | Disproportionating enzyme 1 from Arabidopsis - beta cyclodextrin soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
5N62
 
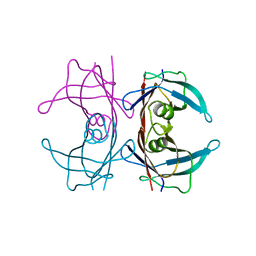 | | Human TTR crystals soaked in manganese chloride. | | Descriptor: | MANGANESE (II) ION, Transthyretin | | Authors: | Ciccone, L, Savko, M, Shepard, W, Stura, E.A. | | Deposit date: | 2017-02-14 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Copper mediated amyloid-beta binding to Transthyretin.
Sci Rep, 8, 2018
|
|
5CU3
 
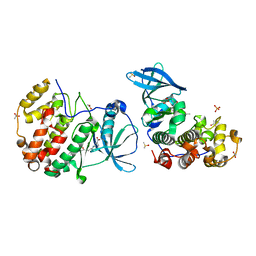 | | Crystal structure of CK2alpha bound to CAM4066 | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, DIMETHYL SULFOXIDE, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
5NB8
 
 | | Structure of vWC domain from CCN3 | | Descriptor: | GLYCEROL, IMIDAZOLE, Protein NOV homolog | | Authors: | Xu, E.-R, Hyvonen, M. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analyses of von Willebrand factor C domains of collagen 2A and CCN3 reveal an alternative mode of binding to bone morphogenetic protein-2.
J. Biol. Chem., 292, 2017
|
|
5CV1
 
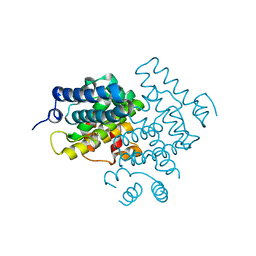 | | C. elegans PGL-1 Dimerization Domain | | Descriptor: | P granule abnormality protein 1 | | Authors: | Aoki, S.T, Bingman, C.A, Wickens, M, Kimble, J.E. | | Deposit date: | 2015-07-25 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.599 Å) | | Cite: | PGL germ granule assembly protein is a base-specific, single-stranded RNase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7CY5
 
 | | Crystal Structure of CMD1 in complex with vitamin C | | Descriptor: | ASCORBIC ACID, CITRIC ACID, FE (III) ION, ... | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
5NCQ
 
 | | Structure of the (SR) Ca2+-ATPase bound to a Tetrahydrocarbazole and TNP-ATP | | Descriptor: | (1~{S})-~{N}-[(4-bromophenyl)methyl]-7-(trifluoromethyloxy)-2,3,4,9-tetrahydro-1~{H}-carbazol-1-amine, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, POTASSIUM ION, ... | | Authors: | Bublitz, M, Kjellerup, L, O'Hanlon Cohrt, K, Gordon, S, Mortensen, A.L, Clausen, J.D, Pallin, D, Hansen, J.B, Brown, W.D, Fuglsang, A, Winther, A.-M.L. | | Deposit date: | 2017-03-06 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Tetrahydrocarbazoles are a novel class of potent P-type ATPase inhibitors with antifungal activity.
PLoS ONE, 13, 2018
|
|
7CXN
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
5ND8
 
 | | Hibernating ribosome from Staphylococcus aureus (Unrotated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Khusainov, I, Vicens, Q, Ayupov, R, Usachev, K, Myasnikov, A, Simonetti, A, Validov, S, Kieffer, B, Yusupova, G, Yusupov, M, Hashem, Y. | | Deposit date: | 2017-03-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF.
EMBO J., 36, 2017
|
|
7D0G
 
 | | Cryo-EM structure of a pre-catalytic group II intron | | Descriptor: | Group II intron-encoded protein LtrA, RNA (714-MER) | | Authors: | Liu, N, Dong, X.L, Hu, C.X, Zeng, J.W, Wang, J.W, Wang, J, Wang, H.W, Belfort, M. | | Deposit date: | 2020-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Exon and protein positioning in a pre-catalytic group II intron RNP primed for splicing.
Nucleic Acids Res., 48, 2020
|
|
5CYL
 
 | |
5N0Z
 
 | | hPAD4 crystal complex with AFM-41a | | Descriptor: | 2-ethyl-~{N}-[(1~{S})-4-(2-fluoranylethanimidoylamino)-1-(4-methoxy-1-methyl-benzimidazol-2-yl)butyl]-3-oxidanylidene-1~{H}-isoindole-4-carboxamide, CALCIUM ION, Protein-arginine deiminase type-4, ... | | Authors: | Beaumont, E, Kerry, P, Thompson, P, Muth, A, Subramanian, V, Nagar, M, Srinath, H, Clancy, K, Parelkar, S. | | Deposit date: | 2017-02-03 | | Release date: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Development of a Selective Inhibitor of Protein Arginine Deiminase 2.
J. Med. Chem., 60, 2017
|
|
5N1B
 
 | | hPAD4 crystal complex with AFM-14a | | Descriptor: | CALCIUM ION, Protein-arginine deiminase type-4, SULFATE ION, ... | | Authors: | Beaumont, E, Kerry, P, Thompson, P, Muth, A, Subramanian, V, Nagar, M, Srinath, H, Clancy, K, Parelkar, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-24 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of a Selective Inhibitor of Protein Arginine Deiminase 2.
J. Med. Chem., 60, 2017
|
|
5NIS
 
 | |
5DDZ
 
 | | Crystal structure of the RTA-c10-P2 complex | | Descriptor: | 60S acidic ribosomal protein P2, Ricin | | Authors: | Zhu, Y, Fan, X, Wang, C, Niu, L, Li, X, Teng, M. | | Deposit date: | 2015-08-25 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the interaction of the ribosomal P stalk protein P2 with a type II ribosome-inactivating protein ricin
Sci Rep, 6, 2016
|
|
1UCO
 
 | | HEN EGG-WHITE LYSOZYME, LOW HUMIDITY FORM | | Descriptor: | LYSOZYME | | Authors: | Nagendra, H.G, Sudarsanakumar, C, Vijayan, M. | | Deposit date: | 1995-12-31 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An X-ray analysis of native monoclinic lysozyme. A case study on the reliability of refined protein structures and a comparison with the low-humidity form in relation to mobility and enzyme action.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
5NJA
 
 | | E. coli Microcin-processing metalloprotease TldD/E with angiotensin analogue bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HIS-PRO-PHE, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5N4J
 
 | |
7CY3
 
 | |
7D0I
 
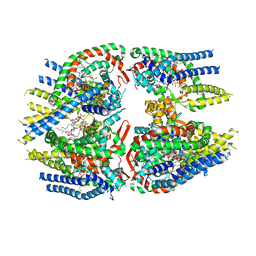 | | Cryo-EM structure of Schizosaccharomyces pombe Atg9 | | Descriptor: | Autophagy-related protein 9, Lauryl Maltose Neopentyl Glycol | | Authors: | Matoba, K, Tsutsumi, A, Kikkawa, M, Noda, N.N. | | Deposit date: | 2020-09-10 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7CY9
 
 | |
7D60
 
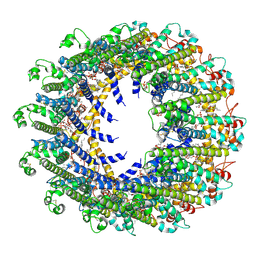 | | Cryo-EM Structure of human CALHM5 in the presence of rubidium red | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, Calcium homeostasis modulator protein 5 | | Authors: | Liu, J, Guan, F.H, Wu, J, Wan, F.T, Lei, M, Ye, S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Cryo-EM structures of human calcium homeostasis modulator 5.
Cell Discov, 6, 2020
|
|
7D7M
 
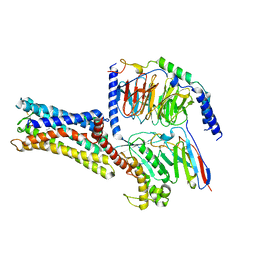 | | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Nojima, S, Fujita, Y, Kimura, T.K, Nomura, N, Suno, R, Morimoto, K, Yamamoto, M, Noda, T, Iwata, S, Shigematsu, H, Kobayashi, T. | | Deposit date: | 2020-10-05 | | Release date: | 2020-11-18 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein.
Structure, 29, 2021
|
|
7D65
 
 | | Cryo-EM Structure of human CALHM5 in the presence of Ca2+ | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, Calcium homeostasis modulator protein 5 | | Authors: | Liu, J, Guan, F.H, Wu, J, Wan, F.T, Lei, M, Ye, S. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structures of human calcium homeostasis modulator 5.
Cell Discov, 6, 2020
|
|
7D7U
 
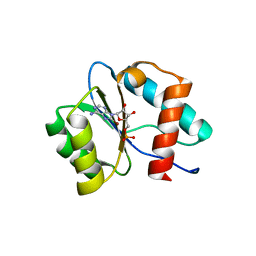 | | Crystal structure of Ago2 MID domain in complex with 8-Br-adenosin-5'-monophosphate | | Descriptor: | 8-BROMO-ADENOSINE-5'-MONOPHOSPHATE, Protein argonaute-2 | | Authors: | Suzuki, M, Takahashi, Y, Saito, J, Miyagi, H, Shinohara, F. | | Deposit date: | 2020-10-06 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | siRNA potency enhancement via chemical modifications of nucleotide bases at the 5'-end of the siRNA guide strand.
Rna, 27, 2021
|
|
