6GSN
 
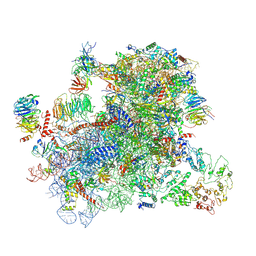 | |
6GSM
 
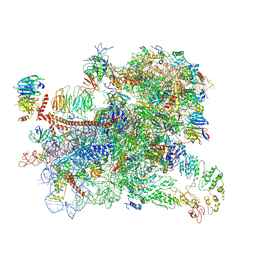 | | Structure of a partial yeast 48S preinitiation complex in open conformation. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Llacer, J.L, Hussain, T, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2018-06-14 | | Release date: | 2019-07-31 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (5.15 Å) | | Cite: | Large-scale movement of eIF3 domains during translation initiation modulate start codon selection.
Nucleic Acids Res., 2021
|
|
9GUI
 
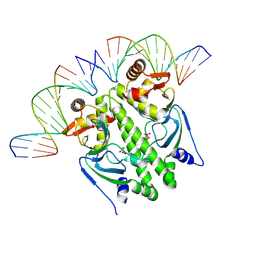 | | Crystal structure of transcription factor NtcA from Synechococcus elongatus in complex with its target DNA. | | Descriptor: | 2-OXOGLUTARIC ACID, DNA (5'-D(P*AP*GP*CP*TP*GP*AP*TP*AP*CP*AP*TP*AP*AP*AP*AP*AP*T)-3'), DNA (5'-D(P*AP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*TP*C)-3'), ... | | Authors: | Llacer, J.L, Forcada-Nadal, A, Rubio, V. | | Deposit date: | 2024-09-19 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the cyanobacterial nitrogen regulators NtcA and PipX complexed to DNA shed light on DNA binding by NtcA and implicate PipX in the recruitment of RNA polymerase.
Nucleic Acids Res., 53, 2025
|
|
9GQU
 
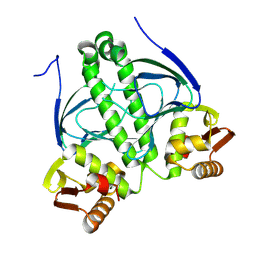 | |
9GUH
 
 | |
9GUJ
 
 | | Crystal structure of transcription factor NtcA from Synechococcus elongatus in complex with its transcriptional co- activator PipX and its target DNA (Crystal II) | | Descriptor: | 2-OXOGLUTARIC ACID, DNA, Global nitrogen regulator, ... | | Authors: | Llacer, J.L, Forcada-Nadal, A, Rubio, V. | | Deposit date: | 2024-09-19 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Structures of the cyanobacterial nitrogen regulators NtcA and PipX complexed to DNA shed light on DNA binding by NtcA and implicate PipX in the recruitment of RNA polymerase.
Nucleic Acids Res., 53, 2025
|
|
9GUG
 
 | |
2XKO
 
 | |
2XHK
 
 | |
2XKP
 
 | |
2XG8
 
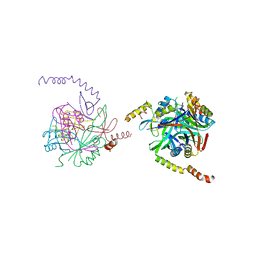 | |
6FYX
 
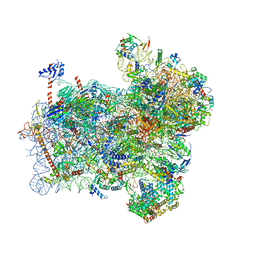 | | Structure of a partial yeast 48S preinitiation complex with eIF5 N-terminal domain (model C1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Llacer, J.L, Hussain, T, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2018-03-12 | | Release date: | 2018-12-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition.
Elife, 7, 2018
|
|
6FYY
 
 | | Structure of a partial yeast 48S preinitiation complex with eIF5 N-terminal domain (model C2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Llacer, J.L, Hussain, T, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2018-03-12 | | Release date: | 2018-12-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition.
Elife, 7, 2018
|
|
2V5H
 
 | | Controlling the storage of nitrogen as arginine: the complex of PII and acetylglutamate kinase from Synechococcus elongatus PCC 7942 | | Descriptor: | ACETYLGLUTAMATE KINASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Llacer, J.L, Marco-Marin, C, Gil-Ortiz, F, Fita, I, Rubio, V. | | Deposit date: | 2007-07-04 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of the Complex of Pii and Acetylglutamate Kinase Reveals How Pii Controls the Storage of Nitrogen as Arginine.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2XGX
 
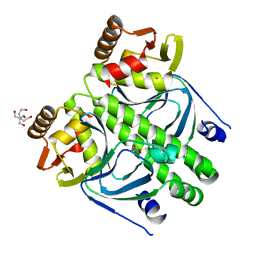 | | Crystal structure of transcription factor NtcA from Synechococcus elongatus (mercury derivative) | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLOBAL NITROGEN REGULATOR, ... | | Authors: | Llacer, J.L, Castells, M.A, Rubio, V. | | Deposit date: | 2010-06-08 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Regulation of Ntca-Dependent Transcription by Proteins Pipx and Pii.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2JJ4
 
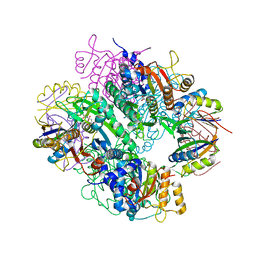 | | The complex of PII and acetylglutamate kinase from Synechococcus elongatus PCC7942 | | Descriptor: | ACETYLGLUTAMATE KINASE, N-ACETYL-L-GLUTAMATE, NITROGEN REGULATORY PROTEIN P-II | | Authors: | Llacer, J.L, Marco-Marin, C, Gil-Ortiz, F, Fita, I, Rubio, V. | | Deposit date: | 2007-07-04 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | The Crystal Structure of the Complex of Pii and Acetylglutamate Kinase Reveals How Pii Controls the Storage of Nitrogen as Arginine
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
3JAP
 
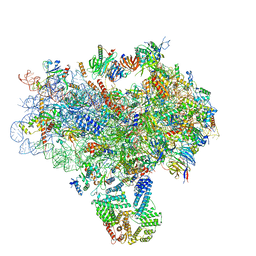 | | Structure of a partial yeast 48S preinitiation complex in closed conformation | | Descriptor: | 18S rRNA, MAGNESIUM ION, METHIONINE, ... | | Authors: | Llacer, J.L, Hussain, T, Ramakrishnan, V. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex.
Mol.Cell, 59, 2015
|
|
3JAM
 
 | | CryoEM structure of 40S-eIF1A-eIF1 complex from yeast | | Descriptor: | 18S rRNA, MAGNESIUM ION, RACK1, ... | | Authors: | Llacer, J.L, Hussain, T, Ramakrishnan, V. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex.
Mol.Cell, 59, 2015
|
|
6QG1
 
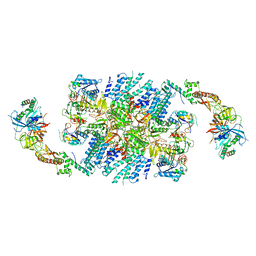 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model 2) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6QG2
 
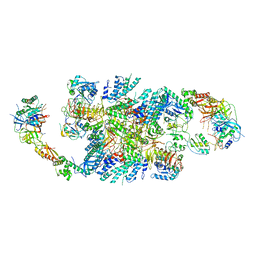 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model A) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6QG6
 
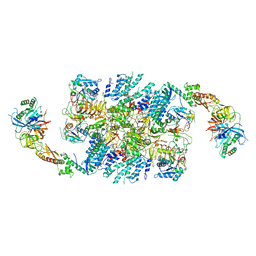 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model D) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6QG3
 
 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model B) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6QG5
 
 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model C) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6QG0
 
 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model 1) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
8P9Y
 
 | | SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | Deposit date: | 2023-06-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
