1AZV
 
 | | FAMILIAL ALS MUTANT G37R CUZNSOD (HUMAN) | | Descriptor: | COPPER (II) ION, COPPER/ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Liu, H, Pellegrini, M, Nersissian, A.M, Gralla, E.B, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1997-11-21 | | Release date: | 1998-02-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Subunit asymmetry in the three-dimensional structure of a human CuZnSOD mutant found in familial amyotrophic lateral sclerosis.
Protein Sci., 7, 1998
|
|
4O9R
 
 | | Human Smoothened Receptor structure in complex with cyclopamine | | Descriptor: | Cyclopamine, Smoothened homolog/Soluble cytochrome b562 chimeric protein | | Authors: | Wang, C, Weierstall, U, James, D, White, T.A, Wang, D, Liu, W, Spence, J.C.H, Doak, R.B, Nelson, G, Fromme, P, Fromme, R, Grotjohann, I, Kupitz, C, Zatsepin, N.A, Liu, H, Basu, S, Wacker, D, Han, G.W, Katritch, V, Boutet, S, Messerschmidt, M, Willams, G.J, Koglin, J.E, Seibert, M.M, Klinker, M, Gati, C, Shoeman, R.L, Barty, A, Chapman, H.N, Kirian, R.A, Beyerlein, K.R, Stevens, R.C, Li, D, Shah, S.T.A, Howe, N, Caffrey, M, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography.
Nat Commun, 5, 2014
|
|
4AVY
 
 | | The AEROPATH project and Pseudomonas aeruginosa high-throughput crystallographic studies for assessment of potential targets in early stage drug discovery. | | Descriptor: | PROBABLE SHORT-CHAIN DEHYDROGENASE | | Authors: | Moynie, L, McMahon, S.A, Alphey, M.S, Liu, H, Duthie, F, Naismith, J.H. | | Deposit date: | 2012-05-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4B79
 
 | | THE AEROPATH PROJECT AND PSEUDOMONAS AERUGINOSA HIGH-THROUGHPUT CRYSTALLOGRAPHIC STUDIES FOR ASSESSMENT OF POTENTIAL TARGETS IN EARLY STAGE DRUG DISCOVERY. | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROBABLE SHORT-CHAIN DEHYDROGENASE | | Authors: | Moynie, L, McMahon, S.A, Alphey, M.S, Liu, H, Duthie, F, Naismith, J.H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4AY9
 
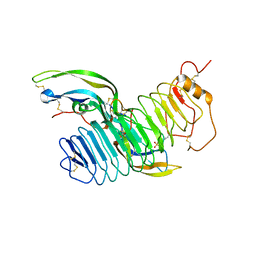 | | Structure of follicle-stimulating hormone in complex with the entire ectodomain of its receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FOLLICLE-STIMULATING HORMONE RECEPTOR, FOLLITROPIN SUBUNIT BETA, ... | | Authors: | Jiang, X, Liu, H, Chen, X, He, X. | | Deposit date: | 2012-06-19 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Follicle-Stimulating Hormone in Complex with the Entire Ectodomain of its Receptor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8JP2
 
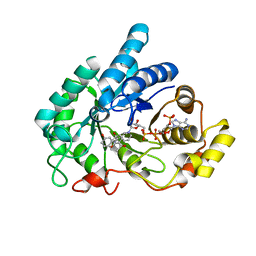 | | Crystal structure of AKR1C1 in complex with DFV | | Descriptor: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X.H, Liu, H, Yao, Z.Q, Zhang, L.P. | | Deposit date: | 2023-06-10 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of AKR1Cs by liquiritigenin and the structural basis.
Chem.Biol.Interact., 385, 2023
|
|
8JP1
 
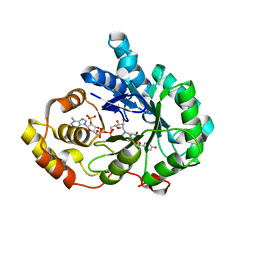 | | Crystal structure of AKR1C3 in complex with DFV | | Descriptor: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X.H, Liu, H, Yao, Z.Q, Zhang, L.P. | | Deposit date: | 2023-06-10 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of AKR1Cs by liquiritigenin and the structural basis.
Chem.Biol.Interact., 385, 2023
|
|
8K93
 
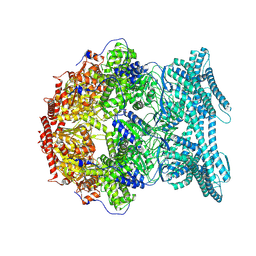 | | CryoEM structure of LonC protease S582A open hexamer with lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K97
 
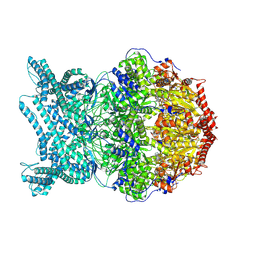 | | CryoEM structure of LonC protease hexamer with Bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, PHOSPHATE ION, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K8Y
 
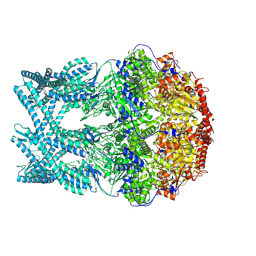 | | CryoEM structure of LonC heptamer in presence of AGS | | Descriptor: | Monothiophosphate, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K8X
 
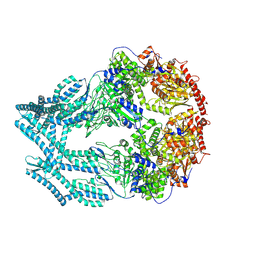 | | CryoEM of LonC open pentamer, apo state | | Descriptor: | endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K96
 
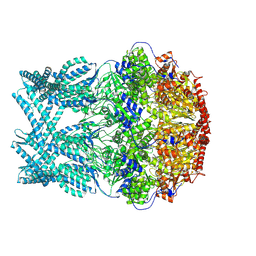 | | CryoEM structure of LonC protease hepatmer with Bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, PHOSPHATE ION, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K92
 
 | | CryoEM structure of LonC S582A hexamer with Lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K8V
 
 | | CryoEM structure of LonC protease hepatmer, apo state | | Descriptor: | PHOSPHATE ION, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K90
 
 | | CryoEM structure of LonC protease open pentamer in presence of AGS | | Descriptor: | Monothiophosphate, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K8Z
 
 | | CryoEM structure of LonC protease hexamer in presence of AGS | | Descriptor: | Monothiophosphate, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K95
 
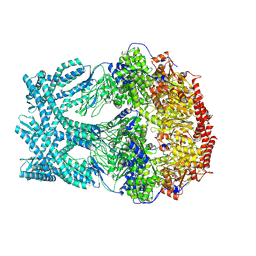 | | CryoEM structure of LonC protease open Hexamer, AGS | | Descriptor: | Monothiophosphate, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K8W
 
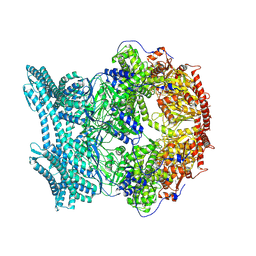 | | CryoEM structure of LonC protease open hexamer, apo state | | Descriptor: | endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K91
 
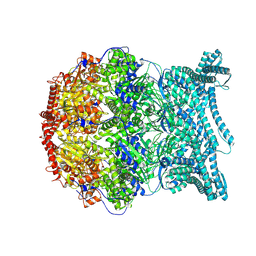 | | CryoEM structure of LonC S582A hepatmer with Lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K94
 
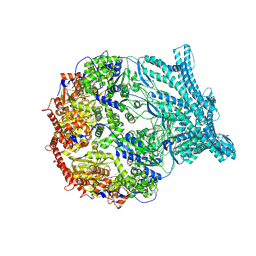 | | CryoEM structure of LonC protease S582A open pentamer with lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
1ORR
 
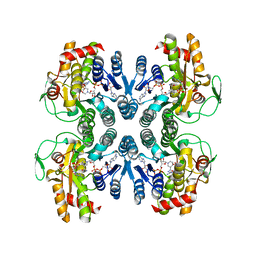 | |
2IVY
 
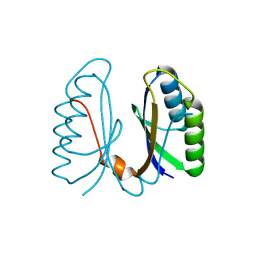 | | Crystal structure of hypothetical protein sso1404 from Sulfolobus solfataricus P2 | | Descriptor: | HYPOTHETICAL PROTEIN SSO1404 | | Authors: | Yan, X, Carter, L.G, Dorward, M, Liu, H, McMahon, S.A, Oke, M, Powers, H, White, M.F, Naismith, J.H. | | Deposit date: | 2006-06-22 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2JG5
 
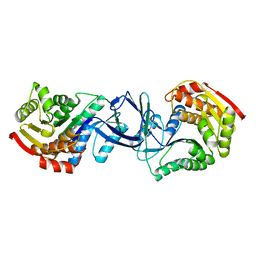 | | CRYSTAL STRUCTURE OF A PUTATIVE PHOSPHOFRUCTOKINASE FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | FRUCTOSE 1-PHOSPHATE KINASE | | Authors: | Yan, X, Carter, L.G, Johnson, K.A, Liu, H, Dorward, M, McMahon, S.A, Oke, M, Powers, H, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2JG6
 
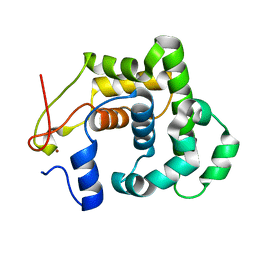 | | CRYSTAL STRUCTURE OF A 3-METHYLADENINE DNA GLYCOSYLASE I FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | DNA-3-METHYLADENINE GLYCOSIDASE, ZINC ION | | Authors: | Yan, X, Carter, L.G, Liu, H, Dorward, M, McMahon, S.A, Johnson, K.A, Oke, M, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
3SJ8
 
 | | Crystal structure of the 3C protease from coxsackievirus A16 | | Descriptor: | 3C protease | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
