4JO8
 
 | | Crystal structure of the activating Ly49H receptor in complex with m157 (G1F strain) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor 8, M157 | | Authors: | Berry, R, Ng, N, Saunders, P.M, Vivian, J.P, Lin, J, Deuss, F.A, Corbett, A.J, Forbes, C.A, Widjaja, J.M, Sullivan, L.C, McAlister, A.D, Perugini, M.A, Call, M.J, Scalzo, A.A, Degli-Esposti, M.A, Coudert, J.D, Beddoe, T, Brooks, A.G, Rossjohn, J. | | Deposit date: | 2013-03-18 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Targeting of a natural killer cell receptor family by a viral immunoevasin
Nat.Immunol., 14, 2013
|
|
4KFO
 
 | | Structure-Based Discovery of Novel Amide-Containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}imidazo[1,2-a]pyridine-6-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Gunzner-Tosteb, J, Liederer, B.M, Ly, J, O'Brien, T, Oh, A, Wang, L, Wang, W, Xiao, Y, Zak, M, Zhao, G, Yuen, P, Bair, K.W. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
4KFN
 
 | | Structure-Based Discovery of Novel Amide-Containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-[4-(piperidin-1-ylsulfonyl)benzyl]-1H-pyrrolo[3,2-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Gunzner-Toste, J, Liederer, B.M, Ly, J, O'Brien, T, Oh, A, Wang, L, Wang, W, Xiao, Y, Zak, M, Zhao, G, Yuen, P, Bair, K.W. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
7ZGP
 
 | |
7ZGR
 
 | |
7ZGQ
 
 | |
6W22
 
 | | ClpA Engaged1 State bound to RepA-GFP (ClpA Focused Refinement) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W24
 
 | | ClpA Engaged2 State bound to RepA-GFP (Focused Classification) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W23
 
 | | ClpA Disengaged State bound to RepA-GFP (Focused Classification) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1PP6
 
 | |
4KJZ
 
 | |
1PP0
 
 | | volvatoxin A2 in monoclinic crystal | | Descriptor: | ACETIC ACID, volvatoxin A2 | | Authors: | Lin, S.-C, Lo, Y.-C, Lin, J.-Y, Liaw, Y.-C. | | Deposit date: | 2003-06-16 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structures and electron micrographs of fungal volvatoxin A2
J.Mol.Biol., 343, 2004
|
|
4GUB
 
 | | Casein Kinase II bound to Inhibitor | | Descriptor: | CHLORIDE ION, Casein kinase II subunit alpha, N-[5-({3-cyano-7-[(1-methyl-1H-imidazol-4-yl)amino]pyrazolo[1,5-a]pyrimidin-5-yl}amino)-2-methylphenyl]acetamide | | Authors: | Larsen, N.A, Dowling, J.E. | | Deposit date: | 2012-08-29 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Potent and selective inhibitors of CK2 kinase.
To be Published
|
|
4J0X
 
 | | Structure of Rrp9 | | Descriptor: | Ribosomal RNA-processing protein 9 | | Authors: | Zhang, L, Lin, J, Ye, K. | | Deposit date: | 2013-01-31 | | Release date: | 2013-06-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural and functional analysis of the U3 snoRNA binding protein Rrp9.
Rna, 19, 2013
|
|
4J0W
 
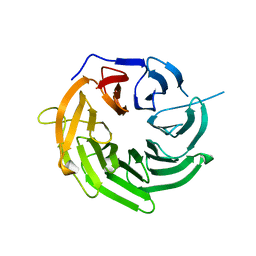 | | Structure of U3-55K | | Descriptor: | U3 small nucleolar RNA-interacting protein 2 | | Authors: | Zhang, L, Lin, J, Ye, K. | | Deposit date: | 2013-01-31 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional analysis of the U3 snoRNA binding protein Rrp9.
Rna, 19, 2013
|
|
3S5B
 
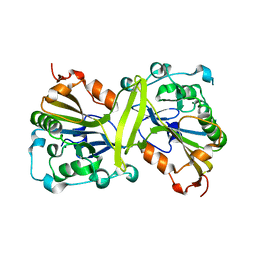 | |
7XOE
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Prefusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
7XOG
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide, ... | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
7VYX
 
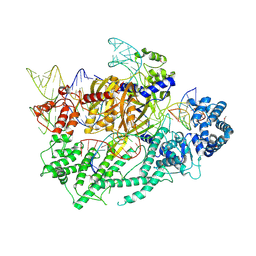 | | Crystal structure of the selenomethionine(SeMet)-derived Cas12c1 (D969A) ternary complex | | Descriptor: | Non-target DNA strand, Selenomethionine (SeMet)-labeled Cas12c1 D969A mutant, Target DNA strand, ... | | Authors: | Zhang, B, Lin, J.Y, Perculija, V, OuYang, S.Y. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-02 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into target DNA recognition and cleavage by the CRISPR-Cas12c1 system
Nucleic Acids Res., 50, 2022
|
|
3DA6
 
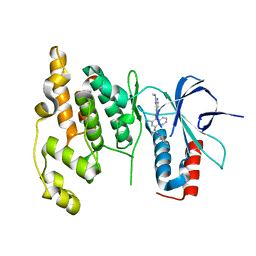 | | Crystal Structure of human JNK3 complexed with N-(3-methyl-4-(3-(2-(methylamino)pyrimidin-4-yl)pyridin-2-yloxy)naphthalen-1-yl)-1H-benzo[d]imidazol-2-amine | | Descriptor: | Mitogen-activated protein kinase 10, N-[3-methyl-4-({3-[2-(methylamino)pyrimidin-4-yl]pyridin-2-yl}oxy)naphthalen-1-yl]-1H-benzimidazol-2-amine | | Authors: | Cee, V.J, Cheng, A.C, Romero, K, Bellon, S, Mohr, C, Whittington, D.A, Bready, J, Caenepeel, S, Coxon, A, Deak, H.L, Hodous, B.L, Kim, J.L, Lin, J, Nguyen, H, Olivieri, P.R, Patel, V.F, Wang, L, Hughes, P, Geuns-Meyer, S. | | Deposit date: | 2008-05-28 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyridyl-pyrimidine benzimidazole derivatives as potent, selective, and orally bioavailable inhibitors of Tie-2 kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4LWW
 
 | | Discovery of Potent and Efficacious Cyanoguanidine-containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-(4-(phenylsulfonyl)benzyl)-1H-pyrrolo[3,2-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynoids, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Oh, A, Wang, W, Zak, M, Wang, L, Yuen, P, Bair, K.W. | | Deposit date: | 2013-07-28 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Discovery of potent and efficacious cyanoguanidine-containing nicotinamide phosphoribosyltransferase (Nampt) inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7UQA
 
 | | Crystal structure of the small Ultra-Red Fluorescent Protein (smURFP) | | Descriptor: | CHLORIDE ION, SODIUM ION, small Ultra-Red Fluorescent Protein (smURFP) | | Authors: | Maiti, A, Buffalo, C.Z, Saurabh, S, Montecinos-Franjola, F, Hachey, J.S, Conlon, W.J, Tran, G.N, Drobizhev, M, Moerner, W.E, Ghosh, P, Matsuo, H, Tsien, R.Y, Lin, J.Y, Rodriguez, E.A. | | Deposit date: | 2022-04-19 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural and photophysical characterization of the small ultra-red fluorescent protein.
Nat Commun, 14, 2023
|
|
4LTS
 
 | | Discovery of Potent and Efficacious Cyanoguanidine-containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 2-cyano-1-pyridin-4-yl-3-(4-{[3-(trifluoromethoxy)phenyl]sulfonyl}benzyl)guanidine, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynoids, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Oh, A, Wang, W, Zak, M, Wang, L, Yuen, P, Bair, K.W. | | Deposit date: | 2013-07-23 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Discovery of potent and efficacious cyanoguanidine-containing nicotinamide phosphoribosyltransferase (Nampt) inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3WTB
 
 | | Crystal structure of Gox0525 | | Descriptor: | Putative oxidoreductase | | Authors: | Yuan, Y.A, Lin, J.P. | | Deposit date: | 2014-04-09 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Gox0525
To be Published
|
|
1GG6
 
 | | CRYSTAL STRUCTURE OF GAMMA CHYMOTRYPSIN WITH N-ACETYL-PHENYLALANINE TRIFLUOROMETHYL KETONE BOUND AT THE ACTIVE SITE | | Descriptor: | 1,1,1-TRIFLUORO-3-ACETAMIDO-4-PHENYL BUTAN-2-ONE(N-ACETYL-L-PHENYLALANYL TRIFLUOROMETHYL KETONE), 1,2-ETHANEDIOL, GAMMA CHYMOTRYPSIN, ... | | Authors: | Neidhart, D, Wei, Y, Cassidy, C, Lin, J, Cleland, W.W, Frey, P.A. | | Deposit date: | 2000-07-31 | | Release date: | 2000-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Correlation of low-barrier hydrogen bonding and oxyanion binding in transition state analogue complexes of chymotrypsin.
Biochemistry, 40, 2001
|
|
