7AYV
 
 | | X-ray crystallographic structure of (6-4)photolyase from Drosophila melanogaster at cryogenic temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, RE11660p, ... | | Authors: | Cellini, A, Wahlgren, W.Y, Henry, L, Westenhoff, S. | | Deposit date: | 2020-11-13 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The three-dimensional structure of Drosophila melanogaster (6-4) photolyase at room temperature.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4S18
 
 | |
6GOH
 
 | | X-ray structure of the adduct formed upon reaction of lysozyme with a Pt(II) complex bearing N,N-pyridylbenzimidazole derivative with an alkylated sulphonate side chain | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2018-06-01 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Exploring the interactions between model proteins and Pd(ii) or Pt(ii) compounds bearing charged N,N-pyridylbenzimidazole bidentate ligands by X-ray crystallography.
Dalton Trans, 47, 2018
|
|
6GOI
 
 | | X-ray structure of the adduct formed upon reaction of lysozyme with a Pd(II) complex bearing N,N-pyridylbenzimidazole derivative with an alkylated triphenylphosphonium cation | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2018-06-01 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Exploring the interactions between model proteins and Pd(ii) or Pt(ii) compounds bearing charged N,N-pyridylbenzimidazole bidentate ligands by X-ray crystallography.
Dalton Trans, 47, 2018
|
|
4UNM
 
 | |
6GH7
 
 | | WILDTYPE CORE-STREPTAVIDIN WITH a conjugated BIOTINYLATED PYRROLIDINE | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[(3~{R})-pyrrolidin-3-yl]pentanamide, Streptavidin | | Authors: | Nodling, A.R, Tsai, Y.H, Luk, L.Y.P, Rizkallah, P, Jin, Y. | | Deposit date: | 2018-05-04 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Reactivity and Selectivity of Iminium Organocatalysis Improved by a Protein Host.
Angew.Chem.Int.Ed.Engl., 57, 2018
|
|
6G5V
 
 | |
6GOK
 
 | | X-ray structure of the adduct formed upon reaction of bovine pancreatic ribonuclease with a Pd(II) complex bearing N,N-pyridylbenzimidazole derivative with an alkylated sulphonate side chain | | Descriptor: | N,N-pyridylbenzimidazole derivative-Pd complex, PALLADIUM ION, Ribonuclease pancreatic | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2018-06-01 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Exploring the interactions between model proteins and Pd(ii) or Pt(ii) compounds bearing charged N,N-pyridylbenzimidazole bidentate ligands by X-ray crystallography.
Dalton Trans, 47, 2018
|
|
6GOB
 
 | | X-ray structure of the adduct formed upon reaction of lysozyme with a Pd(II) complex bearing N,N-pyridylbenzimidazole derivative with an alkylated sulphonate side chain | | Descriptor: | CHLORIDE ION, Lysozyme C, N,N-pyridylbenzimidazole derivative-Pd complex, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2018-06-01 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Exploring the interactions between model proteins and Pd(ii) or Pt(ii) compounds bearing charged N,N-pyridylbenzimidazole bidentate ligands by X-ray crystallography.
Dalton Trans, 47, 2018
|
|
6GOJ
 
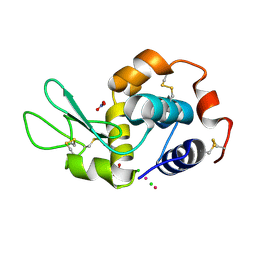 | | X-ray structure of the adduct formed upon reaction of lysozyme with a Pt(II) complex bearing N,N-pyridylbenzimidazole derivative with an alkylated triphenylphosphonium cation | | Descriptor: | CHLORIDE ION, Lysozyme C, NITRATE ION, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2018-06-01 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Exploring the interactions between model proteins and Pd(ii) or Pt(ii) compounds bearing charged N,N-pyridylbenzimidazole bidentate ligands by X-ray crystallography.
Dalton Trans, 47, 2018
|
|
4Z41
 
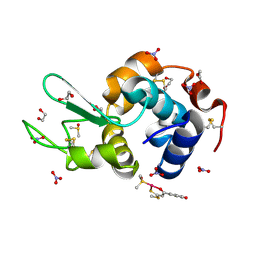 | | X-ray structure of the adduct formed in the reaction between lysozyme and a platinum(II) Compound with a S,O Bidentate Ligand (9a=Chloro-(1-(3'-hydroxy)-3-(methylthio)-3-thioxo-prop-1-en-1-olate-O,S)-(dimethylsulfoxide-S)-platinum(II)) | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-chloranyl-2-[dimethyl(oxidanyl)-{4}-sulfanyl]-4-ethylsulfanyl-1-oxa-3{3}-thia-2{4}-platinacyclohexa-3,5-dien-6-yl]phenol, DIMETHYL SULFOXIDE, ... | | Authors: | Merlino, A. | | Deposit date: | 2015-04-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Platinum(II) Complexes with O,S Bidentate Ligands: Biophysical Characterization, Antiproliferative Activity, and Crystallographic Evidence of Protein Binding.
Inorg.Chem., 54, 2015
|
|
1R5C
 
 | | X-ray structure of the complex of Bovine seminal ribonuclease swapping dimer with d(CpA) | | Descriptor: | 2'-DEOXYCYTIDINE-2'-DEOXYADENOSINE-3',5'-MONOPHOSPHATE, Ribonuclease, seminal | | Authors: | Merlino, A, Vitagliano, L, Sica, F, Zagari, A, Mazzarella, L. | | Deposit date: | 2003-10-10 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Population shift vs induced fit: The case of bovine seminal ribonuclease swapping dimer
Biopolymers, 73, 2004
|
|
6G5Y
 
 | |
9BFY
 
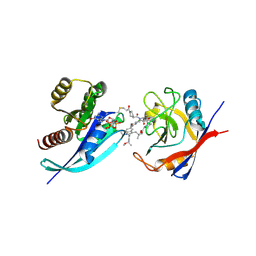 | | Tri-complex of Compound-12, KRAS G12C, and CypA | | Descriptor: | (3R)-N-[(2S)-1-{[(1M,8R,10R,14S,21M)-22-ethyl-4-hydroxy-21-{2-[(1R)-1-methoxyethyl]pyridin-3-yl}-18,18-dimethyl-9,15-dioxo-16-oxa-10,22,28-triazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),2(29),3,5,20,23,26-heptaen-8-yl]amino}-3-methyl-1-oxobutan-2-yl]-N-methyl-1-propanoylpyrrolidine-3-carboxamide (non-preferred name), CHLORIDE ION, GTPase KRas, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-04-18 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Tri-complex of Compound-12, KRAS G12C, and CypA
To be published
|
|
9BG1
 
 | | Tri-complex of Compound-3, KRAS G12V, and CypA | | Descriptor: | (2R)-N-[(1P,7S,9S,13R,20M)-21-ethyl-20-{2-[(1S)-1-methoxyethyl]pyridin-3-yl}-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-methyl-2-(N-methylacetamido)butanamide (non-preferred name), GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-04-18 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Tri-complex of Compound-3, KRAS G12V, and CypA
To be published
|
|
7U4L
 
 | | Crystal structure of human GPX4-U46C in complex with MAC-5576 | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase, thiophene-2-carbaldehyde | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4K
 
 | | Crystal structure of human GPX4-U46C-R152H in complex with ML162 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-(3-chloro-4-methoxyphenyl)-N-[(1R)-2-oxo-2-[(2-phenylethyl)amino]-1-(thiophen-2-yl)ethyl]acetamide, Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Xia, X, Soni, R.K, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4N
 
 | | Crystal structure of human GPX4-U46C in complex with RSL3 | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase, methyl (1S,3R)-2-(chloroacetyl)-1-[4-(methoxycarbonyl)phenyl]-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxylate | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Xia, X, Soni, R.K, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4M
 
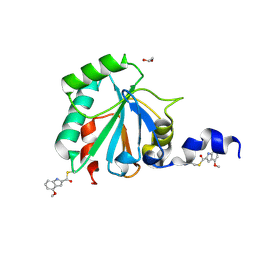 | | Crystal structure of human GPX4-U46C in complex with LOC1886 | | Descriptor: | 1,2-ETHANEDIOL, 4-methoxy-1H-indole-2-carbaldehyde, Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4J
 
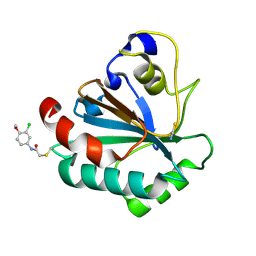 | | Crystal structure of human GPX4-U46C-R152H in complex with TMT10 | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION, ~{N}-(3-chloranyl-4-methoxy-phenyl)ethanamide | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4I
 
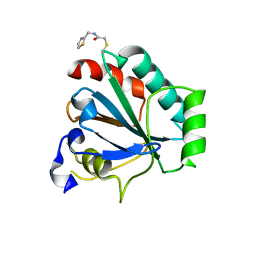 | | Crystal structure of human GPX4-U46C-R152H in complex with CDS9 | | Descriptor: | 2-bromo-N-[(thiophen-2-yl)methyl]acetamide, Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
2I8C
 
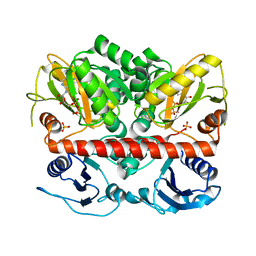 | | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine-D-alanine ligase, MAGNESIUM ION, ... | | Authors: | Liu, S, Chang, J.S, Herberg, J.T, Horng, M, Tomich, P.K, Lin, A.H, Marotti, K.R. | | Deposit date: | 2006-09-01 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4NWF
 
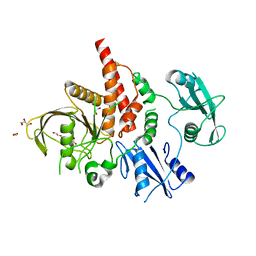 | | Crystal structure of the tyrosine phosphatase SHP-2 with N308D mutation | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Qiu, W, Lin, A, Hutchinson, A, Romanov, V, Ruzanov, M, Thompson, C, Lam, K, Kisselman, G, Battalie, K, Chirgadze, N.Y. | | Deposit date: | 2013-12-06 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the tyrosine phosphatase SHP-2 with N308D mutation
To be Published
|
|
2I80
 
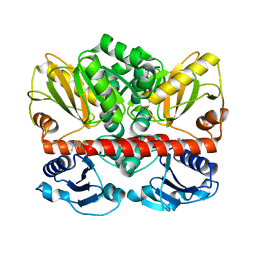 | | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies | | Descriptor: | 3-CHLORO-2,2-DIMETHYL-N-[4-(TRIFLUOROMETHYL)PHENYL]PROPANAMIDE, D-alanine-D-alanine ligase | | Authors: | Liu, S, Chang, J.S, Herberg, J.T, Horng, M.-M, Tomich, P.K, Lin, A.H, Marotti, K.R. | | Deposit date: | 2006-08-31 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4NWG
 
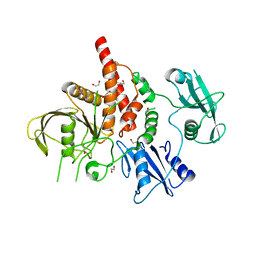 | | Crystal structure of the tyrosine phosphatase SHP-2 with E139D mutation | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Qiu, W, Lin, A, Hutchinson, A, Romanov, V, Ruzanov, M, Thompson, C, Lam, K, Kisselman, G, Battalie, K, Chirgadze, N.Y. | | Deposit date: | 2013-12-06 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the tyrosine phosphatase SHP-2 with E139D mutation
To be Published
|
|
