6PWY
 
 | | Structure of C. elegans ZK177.8, SAMHD1 ortholog | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lim, C.S, Maehigashi, T, Wade, L.R, Bowen, N, Knecht, K, Xiong, Y, Kim, B. | | Deposit date: | 2019-07-24 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | ZK177.8 of Caenorhabditis elegans:
Aicardi-Goutieres Syndrome SAMHD1 Ortholog
To Be Published
|
|
7WVF
 
 | | ectoTLR3-mAb12-poly(I:C) complex | | Descriptor: | RNA (46-MER), Toll-like receptor 3, mAb12 | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-10 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
7WVJ
 
 | | NT-mut(K117D,K139D,K145D) TLR3 -poly I:C complex | | Descriptor: | RNA (46-MER), Toll-like receptor 3 | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-10 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
7WV5
 
 | | ectoTLR3-poly(I:C) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RNA (46-MER), ... | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-09 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
7WV4
 
 | | ectoTLR3-poly(I:C) cluster | | Descriptor: | RNA (80-MER), Toll-like receptor 3 | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-09 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
7WVE
 
 | | CT-mut (D523K,D524K,E527K) TLR3-poly(I:C) complex | | Descriptor: | RNA (46-MER), Toll-like receptor 3 | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-10 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
7WV3
 
 | | Toll-like receptor3 linear cluster | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RNA (80-MER), ... | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-09 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
6X1G
 
 | |
6X1H
 
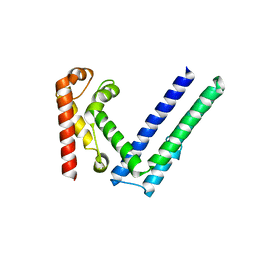 | |
6UPU
 
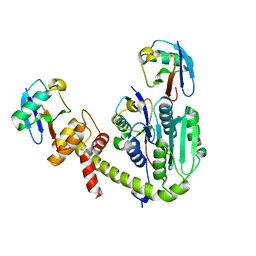 | |
6W6W
 
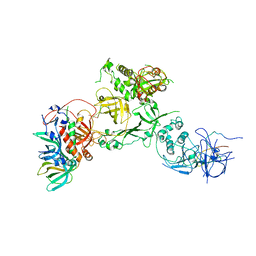 | | Cryo-EM structure of CST bound to telomeric single-stranded DNA | | Descriptor: | CST complex subunit CTC1, CST complex subunit STN1, CST complex subunit TEN1, ... | | Authors: | Lim, C, Barbour, A.T, Zaug, A.J, Goodrich, K.J, McKay, A.E, Wuttke, D.S, Cech, T.R. | | Deposit date: | 2020-03-17 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The structure of human CST reveals a decameric assembly bound to telomeric DNA.
Science, 368, 2020
|
|
1ECL
 
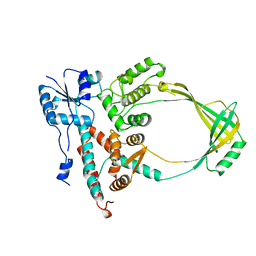 | |
4PN1
 
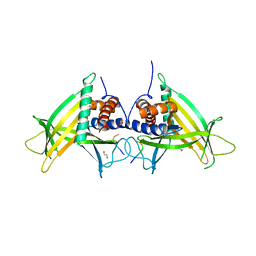 | |
4PN0
 
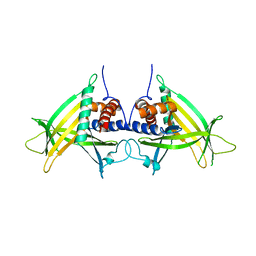 | |
5JNE
 
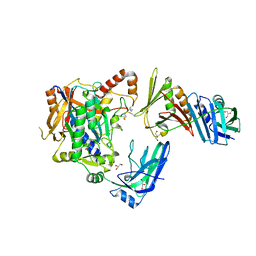 | | E2-SUMO-Siz1 E3-SUMO-PCNA complex | | Descriptor: | E3 SUMO-protein ligase SIZ1,Ubiquitin-like protein SMT3, GLYCEROL, Proliferating cell nuclear antigen, ... | | Authors: | Lima, C.D, Streich Jr, F.C. | | Deposit date: | 2016-04-29 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Capturing a substrate in an activated RING E3/E2-SUMO complex.
Nature, 536, 2016
|
|
5K36
 
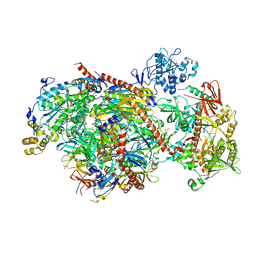 | | Structure of an eleven component nuclear RNA exosome complex bound to RNA | | Descriptor: | Exosome complex component CSL4, Exosome complex component MTR3, Exosome complex component RRP4, ... | | Authors: | Lima, C.D, Zinder, J.C. | | Deposit date: | 2016-05-19 | | Release date: | 2016-11-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nuclear RNA Exosome at 3.1 angstrom Reveals Substrate Specificities, RNA Paths, and Allosteric Inhibition of Rrp44/Dis3.
Mol.Cell, 64, 2016
|
|
6FIT
 
 | | FHIT-TRANSITION STATE ANALOG | | Descriptor: | ADENOSINE MONOTUNGSTATE, FRAGILE HISTIDINE TRIAD PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
6BE2
 
 | |
7KMY
 
 | | Structure of Mtb Lpd bound to 010705 | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Lima, C.D. | | Deposit date: | 2020-11-03 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Whole Cell Active Inhibitors of Mycobacterial Lipoamide Dehydrogenase Afford Selectivity over the Human Enzyme through Tight Binding Interactions.
Acs Infect Dis., 7, 2021
|
|
6BE3
 
 | |
3I2D
 
 | |
5FIT
 
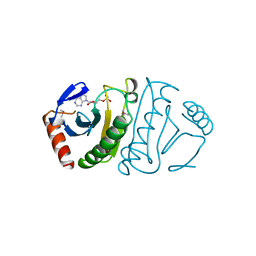 | | FHIT-SUBSTRATE ANALOG | | Descriptor: | FRAGILE HISTIDINE TRIAD PROTEIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
5E6J
 
 | |
3II4
 
 | |
6DG4
 
 | |
