5LRK
 
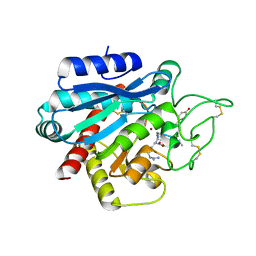 | | Crystal structure of the porcine carboxypeptidase B - Anabaenopeptin F complex | | Descriptor: | Anabaenopeptin F, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Isolation, Co-Crystallization and Structure-Based Characterization of Anabaenopeptins as Highly Potent Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
Sci Rep, 6, 2016
|
|
1VIJ
 
 | | HIV-1 PROTEASE COMPLEXED WITH THE INHIBITOR HOE/BAY 793 HEXAGONAL FORM | | Descriptor: | HIV-1 PROTEASE, N-(1-BENZYL-2,3-DIHYDROXY-4-{3-METHYL-2-[2-(2-METHYL-PROPANE-2-SULFONYLMETHYL)-3-NAPHTHALEN-1-YL-PROPIONYLAMINO]-BUTYRYLAMINO}-5-PHENYL-PENTYL)-3-METHYL-2-[2-(2-METHYL-PROPANE-2-SULFONYLMETHYL)-3-NAPHTHALEN-1-YL-PROPIONYLAMINO]-BUTYRAMIDE | | Authors: | Lange-Savage, G, Berchtold, H, Liesum, A, Hilgenfeld, R. | | Deposit date: | 1997-05-07 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of HOE/BAY 793 complexed to human immunodeficiency virus (HIV-1) protease in two different crystal forms--structure/function relationship and influence of crystal packing.
Eur.J.Biochem., 248, 1997
|
|
1VIK
 
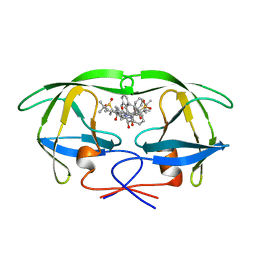 | | HIV-1 PROTEASE COMPLEXED WITH THE INHIBITOR HOE/BAY 793 ORTHORHOMBIC FORM | | Descriptor: | HIV-1 PROTEASE, N-(1-BENZYL-2,3-DIHYDROXY-4-{3-METHYL-2-[2-(2-METHYL-PROPANE-2-SULFONYLMETHYL)-3-NAPHTHALEN-1-YL-PROPIONYLAMINO]-BUTYRYLAMINO}-5-PHENYL-PENTYL)-3-METHYL-2-[2-(2-METHYL-PROPANE-2-SULFONYLMETHYL)-3-NAPHTHALEN-1-YL-PROPIONYLAMINO]-BUTYRAMIDE | | Authors: | Lange-Savage, G, Berchtold, H, Liesum, A, Hilgenfeld, R. | | Deposit date: | 1997-05-07 | | Release date: | 1998-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of HOE/BAY 793 complexed to human immunodeficiency virus (HIV-1) protease in two different crystal forms--structure/function relationship and influence of crystal packing.
Eur.J.Biochem., 248, 1997
|
|
6MV4
 
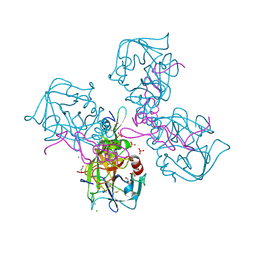 | | CRYSTAL STRUCTURE OF HUMAN COAGULATION FACTOR IXa | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vadivel, K, Schreuder, H.A, Liesum, A, Bajaj, S.P. | | Deposit date: | 2018-10-24 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Sodium-site in serine protease domain of human coagulation factor IXa: evidence from the crystal structure and molecular dynamics simulations study.
J. Thromb. Haemost., 17, 2019
|
|
2BHK
 
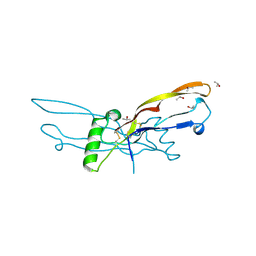 | | Crystal structure of human growth and differentiation factor 5 (GDF5) | | Descriptor: | GROWTH DIFFERENTIATION FACTOR 5, ISOPROPYL ALCOHOL | | Authors: | Schreuder, H, Liesum, A, Pohl, J, Kruse, M, Koyama, M. | | Deposit date: | 2005-01-12 | | Release date: | 2005-03-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Recombinant Human Growth Differentiation Factor 5. Evidence for Interaction of the Type I and Type II Receptor Binding Sites.
Biochem.Biophys.Res.Commun., 329, 2005
|
|
4CIB
 
 | | crystal structure of cathepsin a, complexed with compound 2 | | Descriptor: | 2-(cyclohexylmethyl)propanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Schreuder, H.A, Liesum, A, Kroll, K, Boehnisch, B, Buning, C, Ruf, S, Buning, C, Sadowski, T. | | Deposit date: | 2013-12-06 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of cathepsin A, a novel target for the treatment of cardiovascular diseases.
Biochem. Biophys. Res. Commun., 445, 2014
|
|
4CIA
 
 | | Crystal structure of cathepsin A, complexed with compound 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, LYSOSOMAL PROTECTIVE PROTEIN, ... | | Authors: | Schreuder, H.A, Liesum, A, Kroll, K, Boehnisch, B, Buning, C, Ruf, S, Buning, C, Sadowski, T. | | Deposit date: | 2013-12-06 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of cathepsin A, a novel target for the treatment of cardiovascular diseases.
Biochem. Biophys. Res. Commun., 445, 2014
|
|
4CI9
 
 | | Crystal structure of cathepsin A, apo-structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schreuder, H.A, Liesum, A, Kroll, K, Boehnisch, B, Buning, C, Ruf, S, Buning, C, Sadowski, T. | | Deposit date: | 2013-12-06 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of cathepsin A, a novel target for the treatment of cardiovascular diseases.
Biochem. Biophys. Res. Commun., 445, 2014
|
|
2BMG
 
 | | Crystal structure of factor Xa in complex with 50 | | Descriptor: | 3-[2-(2,4-DICHLOROPHENYL)ETHOXY]-4-METHOXY-N-[(1-PYRIDIN-4-YLPIPERIDIN-4-YL)METHYL]BENZAMIDE, CALCIUM ION, COAGULATION FACTOR X | | Authors: | Schreuder, H, Matter, H, Will, D.W, Nazare, M, Laux, V, Wehner, V, Loenze, P, Liesum, A. | | Deposit date: | 2005-03-14 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Requirements for Factor Xa Inhibition by 3-Oxybenzamides with Neutral P1 Substituents: Combining X-Ray Crystallography, 3D-Qsar and Tailored Scoring Functions
J.Med.Chem., 48, 2005
|
|
1GK0
 
 | | Structure-based prediction of modifications in glutarylamidase to allow single-step enzymatic production of 7-aminocephalosporanic acid from cephalosporin C | | Descriptor: | 1,2-ETHANEDIOL, CEPHALOSPORIN ACYLASE, PHOSPHATE ION | | Authors: | Fritz-Wolf, K, Koller, K.P, Lange, G, Liesum, A, Sauber, K, Schreuder, H, Aretz, W, Kabsch, W. | | Deposit date: | 2001-08-07 | | Release date: | 2002-01-01 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Prediction of Modifications in Glutarylamidase to Allow Single-Step Enzymatic Production of 7-Aminocephalosporanic Acid from Cephalosporin C.
Protein Sci., 11, 2002
|
|
1GK1
 
 | | Structure-based prediction of modifications in glutarylamidase to allow single-step enzymatic production of 7-aminocephalosporanic acid from cephalosporin C | | Descriptor: | CEPHALOSPORIN ACYLASE, GLYCEROL | | Authors: | Fritz-Wolf, K, Koller, K.P, Lange, G, Liesum, A, Sauber, K, Schreuder, H, Aretz, W, Kabsch, W. | | Deposit date: | 2001-08-07 | | Release date: | 2002-01-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Prediction of Modifications in Glutarylamidase to Allow Single-Step Enzymatic Production of 7-Aminocephalosporanic Acid from Cephalosporin C.
Protein Sci., 11, 2002
|
|
1LPK
 
 | | CRYSTAL STRUCTURE OF FXA IN COMPLEX WITH 125. | | Descriptor: | 1-(3-CARBAMIMIDOYL-BENZYL)-1H-INDOLE-2-CARBOXYLIC ACID 3-CARBAMIMIDOYL-BENZYLESTER, Blood coagulation factor Xa, CALCIUM ION | | Authors: | Schreuder, H.A, Loenze, P, Brachvogel, V, Liesum, A. | | Deposit date: | 2002-05-08 | | Release date: | 2003-05-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and Quantitative Structure-Activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa
J.Med.Chem., 45, 2002
|
|
1LQD
 
 | | CRYSTAL STRUCTURE OF FXA IN COMPLEX WITH 45. | | Descriptor: | 1-(3-CARBAMIMIDOYL-BENZYL)-4-METHYL-1H-INDOLE-2-CARBOXYLIC ACID 3,5-DIMETHYL-BENZYLAMIDE, Blood coagulation factor Xa, CALCIUM ION | | Authors: | Schreuder, H.A, Loenze, P, Brachvogel, V, Liesum, A. | | Deposit date: | 2002-05-10 | | Release date: | 2003-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and Quantitative Structure-Activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa
J.Med.Chem., 45, 2002
|
|
1LPG
 
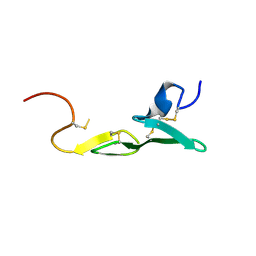 | | CRYSTAL STRUCTURE OF FXA IN COMPLEX WITH 79. | | Descriptor: | Blood coagulation factor Xa, CALCIUM ION, [4-({[5-BENZYLOXY-1-(3-CARBAMIMIDOYL-BENZYL)-1H-INDOLE-2-CARBONYL]-AMINO}-METHYL)-PHENYL]-TRIMETHYL-AMMONIUM | | Authors: | Schreuder, H.A, Brachvogel, V, Liesum, A. | | Deposit date: | 2002-05-08 | | Release date: | 2003-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Quantitative Structure-Activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa.
J.Med.Chem., 45, 2002
|
|
1LPZ
 
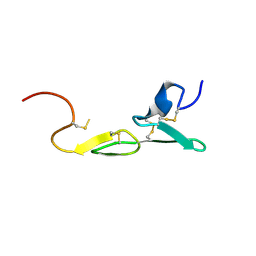 | | CRYSTAL STRUCTURE OF FXA IN COMPLEX WITH 41. | | Descriptor: | 1-(3-carbamimidoylbenzyl)-N-(3,5-dichlorobenzyl)-4-methyl-1H-indole-2-carboxamide, Blood coagulation factor Xa, CALCIUM ION | | Authors: | Schreuder, H.A, Brachvogel, V, Liesum, A. | | Deposit date: | 2002-05-08 | | Release date: | 2003-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Quantitative Structure-Activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa
J.Biol.Chem., 45, 2002
|
|
1LQE
 
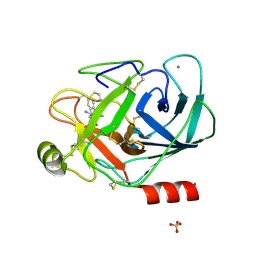 | | CRYSTAL STRUCTURE OF TRYPSIN IN COMPLEX WITH 79. | | Descriptor: | CALCIUM ION, SULFATE ION, TRYPSIN, ... | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2002-05-10 | | Release date: | 2003-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and quantitative structure-activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides
as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa.
J.Med.Chem., 45, 2002
|
|
5LRJ
 
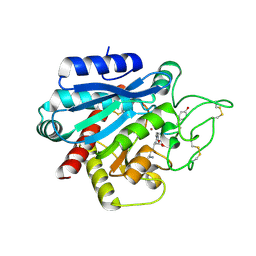 | | Crystal structure of the porcine carboxypeptidase B - Anabaenopeptin C complex | | Descriptor: | Anabaenopeptin C, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isolation, Co-Crystallization and Structure-Based Characterization of Anabaenopeptins as Highly Potent Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
Sci Rep, 6, 2016
|
|
5LRG
 
 | | Crystal structure of the porcine carboxypeptidase B - Anabaenopeptin B complex | | Descriptor: | Anabaenopeptin B, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Isolation, Co-Crystallization and Structure-Based Characterization of Anabaenopeptins as Highly Potent Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
Sci Rep, 6, 2016
|
|
2AER
 
 | | Crystal Structure of Benzamidine-Factor VIIa/Soluble Tissue Factor complex. | | Descriptor: | BENZAMIDINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bajaj, S.P, Schmidt, A.E, Padmanabhan, K, Bajaj, M.S, Liesum, A, Dumas, J, Prevost, D, Schreuder, H. | | Deposit date: | 2005-07-23 | | Release date: | 2006-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | High Resolution Structures of p-Aminobenzamidine- and Benzamidine-VIIa/Soluble Tissue Factor: Unpredicted conformation of the 192-193 peptide bond and mapping of Ca2+, Mg2+, Na+ and Zn2+ sites in factor VIIa
J.Biol.Chem., 281, 2006
|
|
6GB1
 
 | | Crystal structure of the GLP1 receptor ECD with Peptide 11 | | Descriptor: | Glucagon-like peptide 1 receptor, HEXANE-1,6-DIOL, Peptide 11, ... | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2018-04-13 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Dual Glucagon-like Peptide 1 (GLP-1)/Glucagon Receptor Agonists Specifically Optimized for Multidose Formulations.
J. Med. Chem., 61, 2018
|
|
6YG9
 
 | | CRYSTAL STRUCTURE OF HUMAN SERUM ALBUMIN (HSA) IN COMPLEX WITH GN-07. | | Descriptor: | 20-[[(2~{S})-5-[2-[2-[2-[2-[2-[2-(diethylamino)-2-oxidanylidene-ethoxy]ethoxy]ethylamino]-2-oxidanylidene-ethoxy]ethoxy]ethylamino]-1-oxidanyl-1,5-bis(oxidanylidene)pentan-2-yl]amino]-20-oxidanylidene-icosanoic acid, MYRISTIC ACID, Serum albumin | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2020-03-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Triantennary GalNAc Molecular Imaging Probes for Monitoring Hepatocyte Function in a Rat Model of Nonalcoholic Steatohepatitis.
Adv Sci, 7, 2020
|
|
6YAU
 
 | | CRYSTAL STRUCTURE OF ASGPR 1 IN COMPLEX WITH GN-A. | | Descriptor: | 5-[(2~{R},3~{R},4~{R},5~{R},6~{R})-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-~{N}-[3-(propanoylamino)propyl]pentanamide, Asialoglycoprotein receptor 1, CALCIUM ION | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2020-03-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.397 Å) | | Cite: | Triantennary GalNAc Molecular Imaging Probes for Monitoring Hepatocyte Function in a Rat Model of Nonalcoholic Steatohepatitis.
Adv Sci, 7, 2020
|
|
7BKG
 
 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with the tricyclic inhibitor (2) | | Descriptor: | 5,6-dihydro-2-imino-2H,4H-thiazolo(5,4,3-IJ)quinoline, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2021-01-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.326 Å) | | Cite: | Novel Inhibitors of Nicotinamide- N -Methyltransferase for the Treatment of Metabolic Disorders.
Molecules, 26, 2021
|
|
7BLE
 
 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with the tricyclic inhibitor (3) | | Descriptor: | 3-ethyl-1,3-diazatricyclo[6.3.1.0^{4,12}]dodeca-4,6,8(12)-trien-2-imine, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Novel Inhibitors of Nicotinamide- N -Methyltransferase for the Treatment of Metabolic Disorders.
Molecules, 26, 2021
|
|
7NBJ
 
 | |
